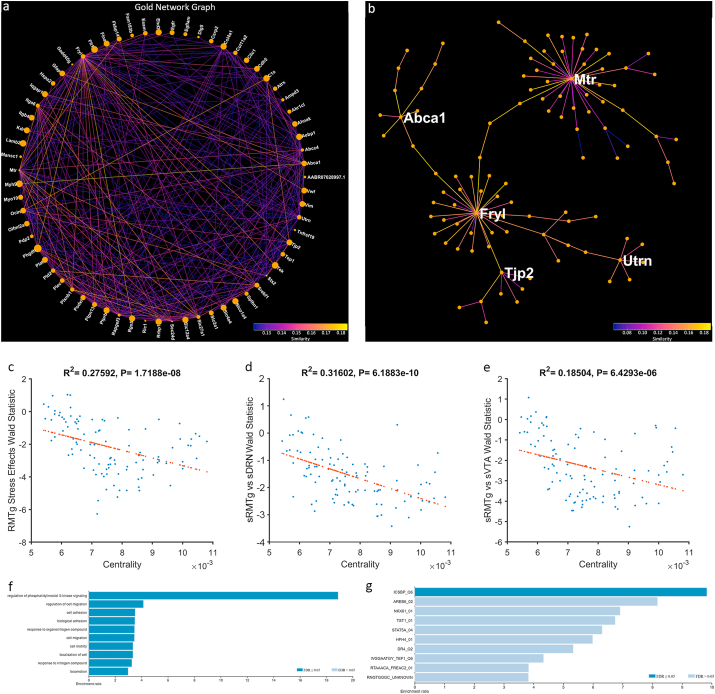
Fig. 7.
The Gold Network. a) Gold Gene Network projected onto a two-dimensional space using a circle layout. Line color represents pairwise gene expression similarity as calculated by WGCNA. Node size represents centrality to the network. b) Gold Network projected as a minimum spanning tree. The most central genes are labeled. c)Genes which are more downregulated with stress in the RMTg pathway are more central to the network (r2 = 0.276, p < 0.001). d) Genes which are less highly expressed in the stressed RMTg than the stressed DRN pathway are more central to the network (r2 = 0.316, p < 0.001). e) Genes which are less highly expressed in the stressed RMTg than the stressed VTA pathway are more central to the network (r2 = 0.185, p < 0.001). f) Overrepresentation analysis of GO: Biological Process of the entire network. g) Overrepresentation analysis of transcription factor targets for the 25 most central genes to the network. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)