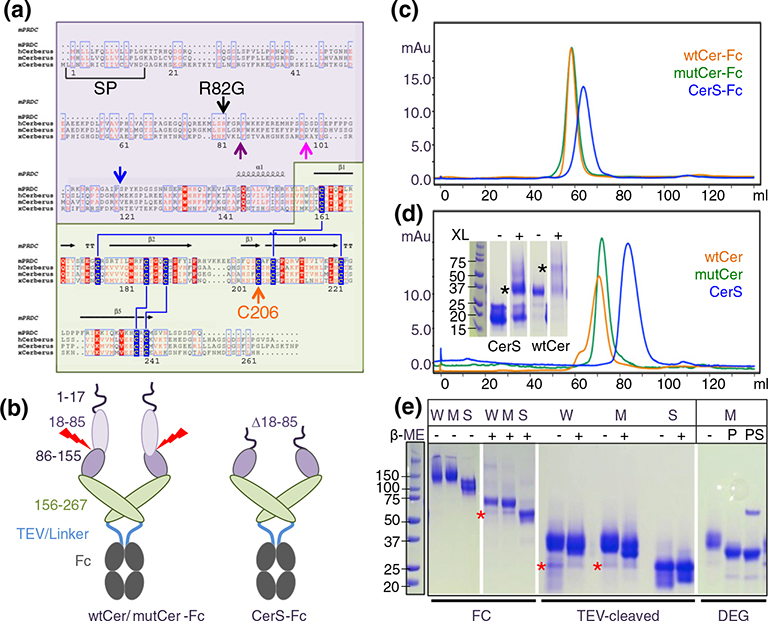
Fig. 1.
Construct design and purification. (a) Multiple sequence alignment of the DAN family proteins PRDC (mouse) and Cerberus (human, mouse, and frog). DAN family proteins are secreted regulators of TGF-β family signaling that have a signal peptide for secretion (SP) and a conserved cystine knot domain (highlighted in green, Cerberus amino acids 156–267). Cerberus has a unique N-terminal region of unknown function (highlighted in purple, Cerberus amino acids 19–155). Cerberus molecules are posttranslationally processed and have a predicted proprotein convertase processing site, marked by the purple arrow for mammalian Cerberus and by a blue arrow for frog Cerberus [29], or a proposed processing site marked by a pink arrow [1]. Cerberus molecules also have an unpaired cysteine (Cys206 in human Cerberus, orange arrow). Arginine 82 and cysteine 206 were mutated for functional analysis. (b) Domain organization of Cerberus and construct design. Cerberus consists of three distinct regions, the cleavable N-terminal region (amino acids 19–85, light purple), the residual N-terminal region (amino acids 86–155, dark purple), and the cystine knot domain (amino acids 156–267, green). Three different constructs were created: Full-length human Cerberus with the wild-type sequence (wtCer-Fc), full-length Cerberus with mutations at arginine 82 and cysteine 206 (mutCer-Fc), and a short form lacking the N-terminal region (amino acids 18–85) and mutated at cysteine 206 (CerS-Fc). Cerberus genes were fused at the C-terminus to human IgG1-Fc via a 22-amino-acid linker containing a TEV cleavage site. (c) Purification of wtCer-Fc, mutCer-Fc, and CerS-Fc expressed in CHO cells. Following two purification steps, molecules migrate as a single, well-defined peak in a size-exclusion chromatographic column. The molecular weight of each protein corresponds to the dimeric species. (d) Purification of wtCer, mutCer, and CerS after removal of the Fc domain. All molecules migrate as a single, well-defined peak in a SEC column. For wtCer and mutCer, the molecular weight corresponds to the dimeric species. CerS elutes at a volume that corresponds to a monomeric form, but it also forms dimers in solution. The inserted SDS-PAGE shows glutaraldehyde cross-linking of CerS. The dimeric CerS is highlighted by the black asterisk. For functional studies, only the main peak fractions were used and fractions corresponding to higher-molecular-weight species were discarded. (e) SDS-PAGE gels of purified proteins. The two left panels show non-reducing (−β-ME) and reducing (+β-ME) SDS-PAGE gels of the Fc fusion forms (W: wtCer-Fc, M: mutCer-Fc, S: CerS-Fc). Expected molecular masses are 57 kDa for wtCer-Fc, 57 kDa for mutCer-Fc, and 49 kDa for CerS-Fc. Higher apparent molecular weights are due to glycosylation. wtCer-Fc and mutCer-Fc have three N-linked glycosylation sites per protomer, two in the Cerberus moiety and one in the Fc moiety. CerS-Fc has two N-linked glycosylation sites per protomer, one in the Cerberus moiety and one in the Fc moiety. Fc fusion constructs form disulfide-linked dimers via the Fc domain. The two right panels show Fc free Cerberus. The molecular weights of the three cleaved Cerberus constructs correspond to a monomeric form under reducing and non-reducing conditions. Deglycosylation with PNGase F alone (P) or with PNGase F and sialidase (PS) reduces the molecular weight of mutant Cerberus to the theoretically expected value. The red star designates processed Cerberus.