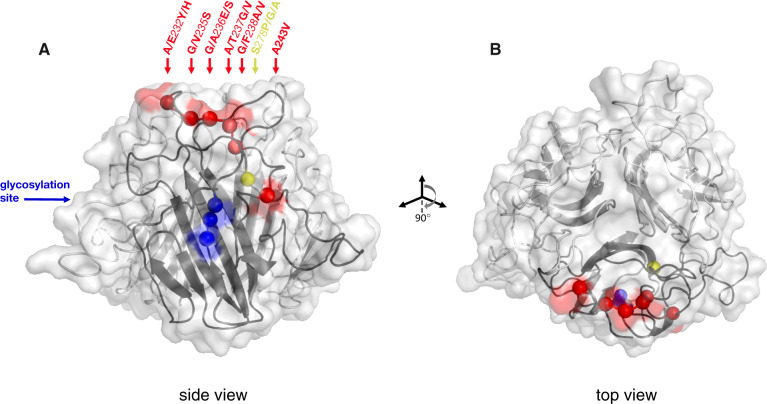
Fig. 4.
(A) Structural model of C-terminal globular domain of Pcl7 homotrimer, based on alignment with BclA. The collagen-like triple-helix domain and the N-terminus (not shown) would descend from the bottom center of the structure shown. The backbone is illustrated as a ribbon diagram with the transparent protein surface overlaid. Balls along the backbone of one of the three identical monomers indicate the locations of polymorphic residues whose variation segregates perfectly by infection phenotype. Balls and any corresponding surface-exposed area are highlighted according to their coordinates in the protein alignment: Red for the cluster of polymorphisms at residues 232–243, yellow for the polymorphism at residue 278, blue for the segregating glycosylation site at residues 308–310. Polymorphisms are named according to the residue(s) of the infectious haplotypes, the coordinate in the alignment, and the residue(s) of the noninfectious haplotypes. All polymorphisms are surface exposed, except residues 238 and 278. (B) The structure has been rotated forward 90° to show the top view.