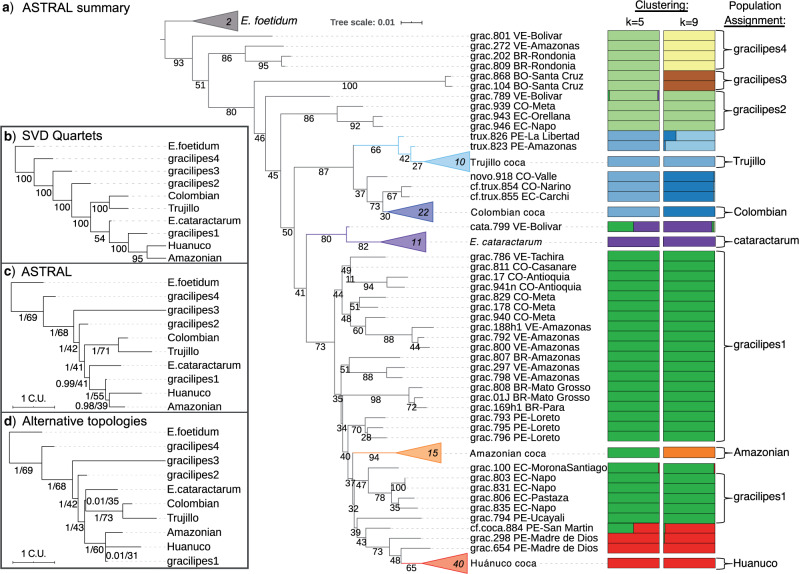
Figure 2.
Phylogeny and genetic clustering of coca varieties and wild relatives. a) ASTRAL-III summary (AST) tree topology with branch lengths optimized using the ML-SNP data set; scale indicates substitutions per site and branch support values denote concordance factors of SNPs; collapsed clades are marked with the number of samples; tip names indicate taxon, sample ID, and geographic provenance indicated by the two letter country code and political subdivision. Probability assignment of individuals to five or nine genetic clusters are indicated in bar plots along with the population assignment of ‘pure’ individuals for species trees and population genetic statistics. b) SVDquartets species tree inferred from SNP data with bootstrap support. c) ASTRAL-III species tree inferred from 424 gene trees with local posterior probability (LPP) and percent quartet support, respectively; branch lengths scaled by coalescent units. d) Constrained ASTRAL-III species trees indicating LPP and percent quartet support for an E. cataractarum 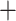 Trujillo/Colombian clade or a Huánuco
Trujillo/Colombian clade or a Huánuco 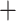 gracilipes1 clade.
gracilipes1 clade.