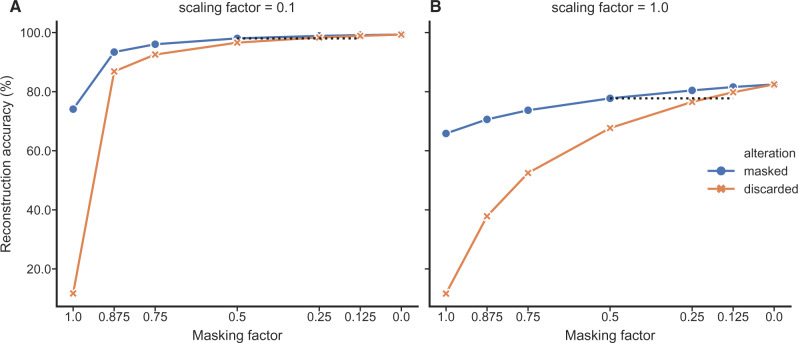
Figure 7.
The accuracy of ancestral rotamer sequence reconstruction from mixed data under RAM55 increases when masked sequences, which lack rotamer states, are not discarded. The x-axis shows the fraction of information available under two scenarios. The dark circles reflect the amount of rotamer information that has been masked (i.e., replaced with amino acids), and the light crosses represent the amount of rotamer sequences that has been replaced with gaps (discarded). Masking half of the rotamer configurations produces accuracies comparable with those obtained by replacing 1/8–1/4 of sequences in the alignments with gaps (see black dashed lines). Reconstructions with ambiguity always outperform discarding an equivalent fraction of amino acid sequences. As before, the shallower tree, A), shows higher overall accuracy.