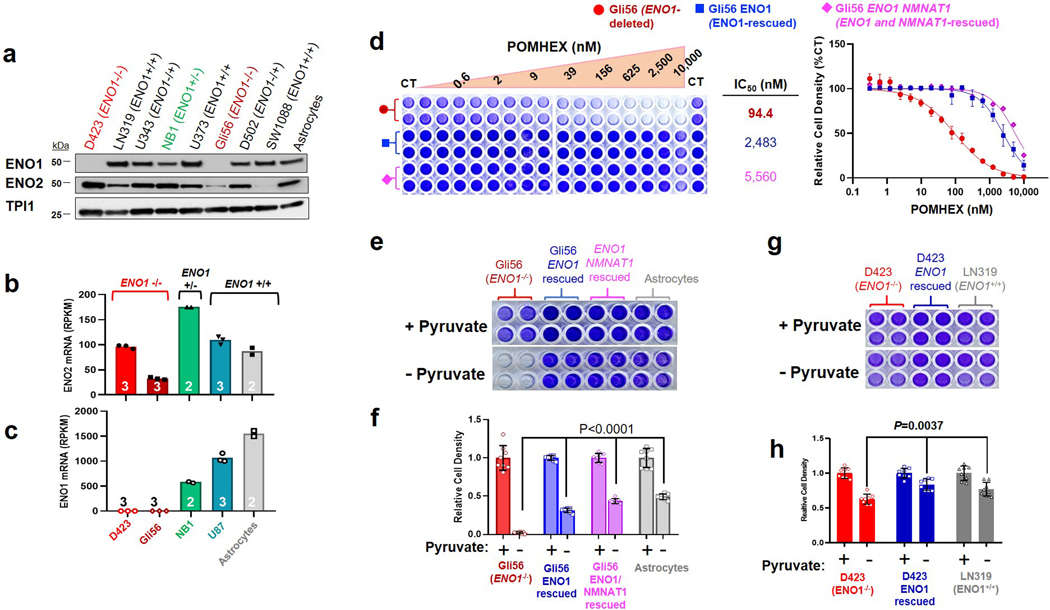
Extended Data Fig. 3. The Gli56 ENO1-homozygously deleted glioma cell line is sensitive to POMHEX and is pyruvate auxotrophic.
Sensitivity to POMHEX was determined in additional ENO1-homozygous deleted cell line, Gli56 (dark red) and its isogenic ENO1-rescued control3,4. a. Absence of ENO1 protein confirmed by western blot in the Gli56 cell line; note levels of ENO2 are lower than in D423 (ENO1−/−), which is confirmed by mRNA. b, c. RNA-seq mRNA expression of ENO1 and ENO2 in cell lines differing in ENO1-deletion status, confirming lower ENO2-expression in Gli56 compared to D423 cell. Each point represent one biological replicate, means are shown, n number is indicated for each cell line. d. Gli56 (dark red) is selectively sensitive to POMHEX compared to its isogenic ENO1-rescued control3 (blue). Another Gli56 rescued cell line (purple), in which ENO1 and a neighboring deleted gene, NMNAT1, are re-expressed (generated for a separate collateral lethality project), also shows resistance to POMHEX. Cell density was visualized by crystal violet after 14 days of growth; fresh media (DMEM, contains 1.25 mM pyruvate) containing Enolase inhibitor was provided every 4 days (N = 4 biological replicates +/− S.EM.). In contrast to the D423 cell line, Gli56 cannot be grown in pyruvate-free DMEM media, likely due to lower level of residual ENO2 expression. e, f. Gli56 ENO1-deleted, but not Gli56 ENO1-rescued cells, can grow in pyruvate-free DMEM. g, h. D423 ENO1-deleted cells can grow in pyruvate-free DMEM media, but slower than D423 ENO1-rescued or LN319 (ENO1-WT) cells. N = 8 biological replicates, mean +/− S.E.M. P value by 2-way ANOVA are indicated.