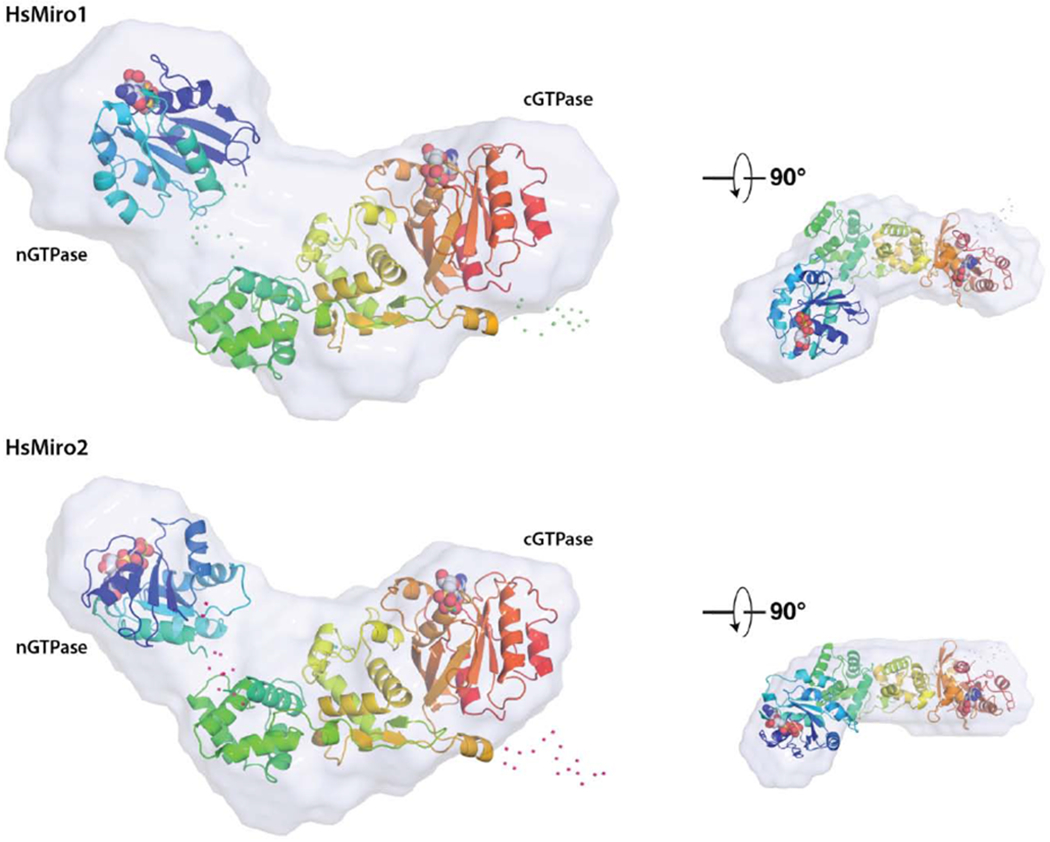
Figure 4. Solution structure of monomeric HsMiro1 and HsMiro2.
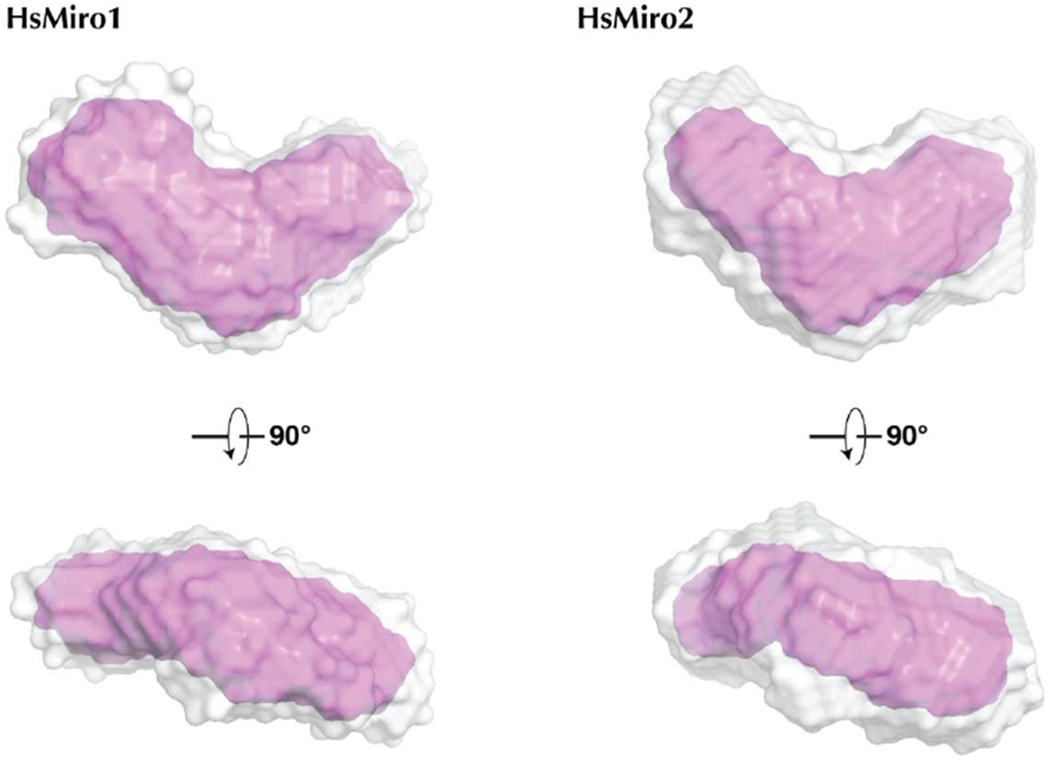
(A) Averaged reconstructions from 10 DAMMIF calculations of the processed SAXS data from HsMiro1 and HsMiro2. The DAMAVER average of the DAMMIF runs is shown in gray and the DAMFILT filtered envelope is shown in pink [61,79]. The dummy atoms used to construct the contours shown were generated with a radius of 4Å.
(B) Representative monomer models generated using BUNCH [64] are overlaid with the corresponding SAXS scattering envelope reconstructions. The two models are aligned with respect to the MiroS domain, with the nGTPase at left in each case. The positions of the nucleotides bound to each of the GTPase domains are shown as spheres. Shown smaller at right are the overlays viewed after a ~90° rotation around the horizontal axis. The scattering envelopes reasonably enclose both the rod-like MiroS and nGTPase domains. Note that while the orientations of MiroS in the two structures are similar, the orientations of the nGTPase domains are different, reflecting the fact that its position cannot be fixed based on this data; their relative dispositions suggest that the nGTPase domains are not tightly associated with the ELM1 domain of MiroS. Dots represent amino acids not observed in the crystal structures and modeled as flexible scattering elements.
