Figure 1:
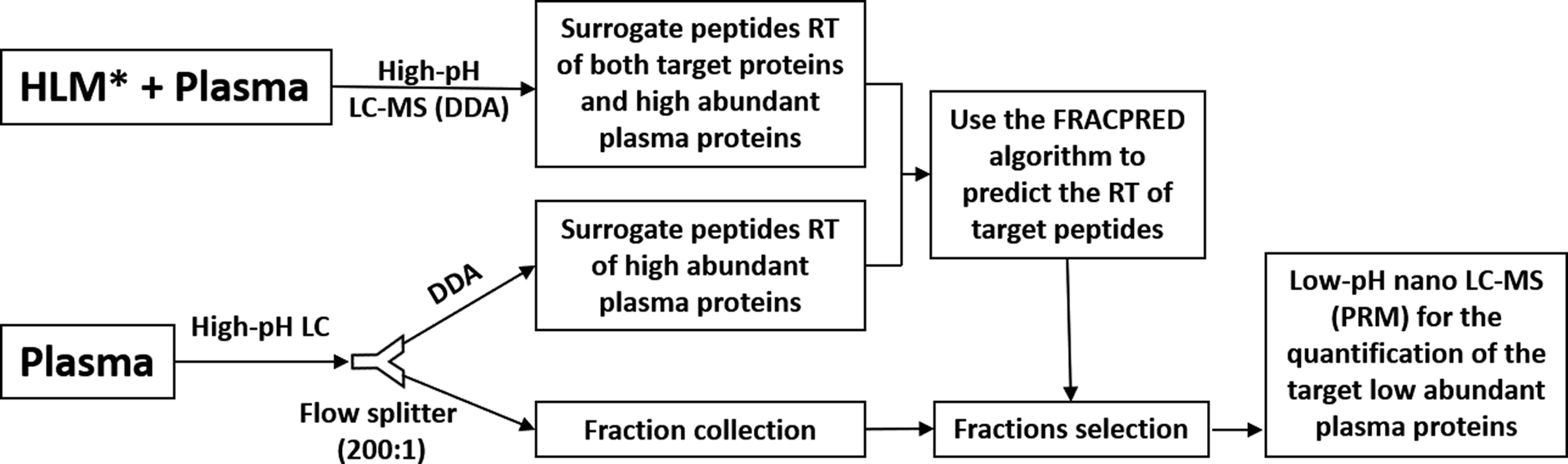
Workflow of the FRACPRED-2D-PRM approach. The digested plasma samples or the mixed plasma and human liver microsomes (HLM) samples are first separated with a high-pH mobile phase on a large diameter C18 column (4.6 mm × 150 mm). A post-column splitter is used to split the flow (1 ml/min) at a 1:200 ratio with 5 μl/min being directed to a mass spectrometer for data-dependent acquisition (DDA) and 995 μl/min for fraction collection at 30 or 15-second intervals. The resulting DDA data are individually searched by the MaxQuant software then imported into the Skyline software to generate a list of peptides and their retention time. The retention time (RT) of target peptides in the plasma samples can be predicted using the FRACPRED algorithm (R package, fracpred), and the corresponding fractions can then be selected for the second dimension low-pH parallel reaction monitoring (PRM) analysis.
*We chose HLM because the targeted proteins are highly expressed in the liver. Other types of tissues could be used depending on the expression levels of the targeted proteins in the tissues.
