Figure 5. Single cell RNA-seq analyses on effects of Hedgehog activation on major types of SMG cells and their paracrine interactions.
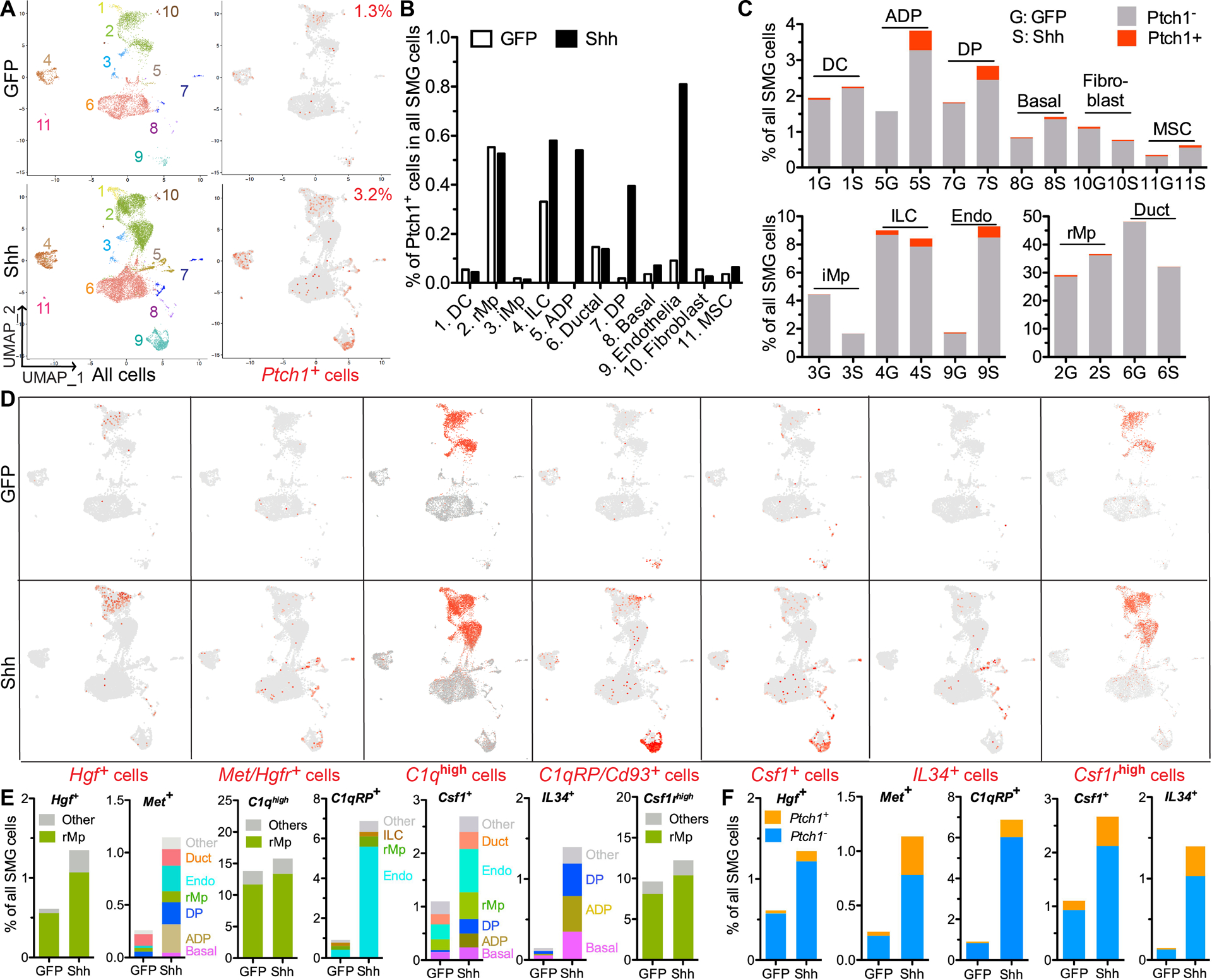
A-C: At 7 days after intra-SMG transfer of GFP or Shh gene, SMG cells were analzyed with sc RNA-seq to explore the identities and percentages of all major types of SMG cells and Ptch1+ Hedgehog-responsive cells based on the expression of corresponding marker genes. The putative identities of clusters are 1) DC: dendritic cells, 2) rMp: resident macrophages, 3) iMp: infiltrating macrophages, 4) ILCs: innate lymphoid cells, 5) ADP: acinar/ductal progenitors, 6) ductal cells, 7) DP: ductal progenitors, 8) Basal: basal/myo-epithelial cells, 9) Endothelia, 10) Fibroblasts, 11) MSC: mesenchymal stem cells.
D-E: In these cell clusters, cells were labeled red for all detectable expression of Hgf, Met/Hgfr, Cd93, Csf1 or IL34 and for high expression (>50th percentile) of all C1q genes (C1qa, C1qb, C1qc) or Csf1r (D). Cells positive for these genes in each cell clusters were quantified as percentages of all SMG cells (E).
F. In cells expressing the above genes except C1q and Csf1r, Ptch1+ and Ptch1− cells were quantified as percentages of all SMG cells.
