Figure 4.
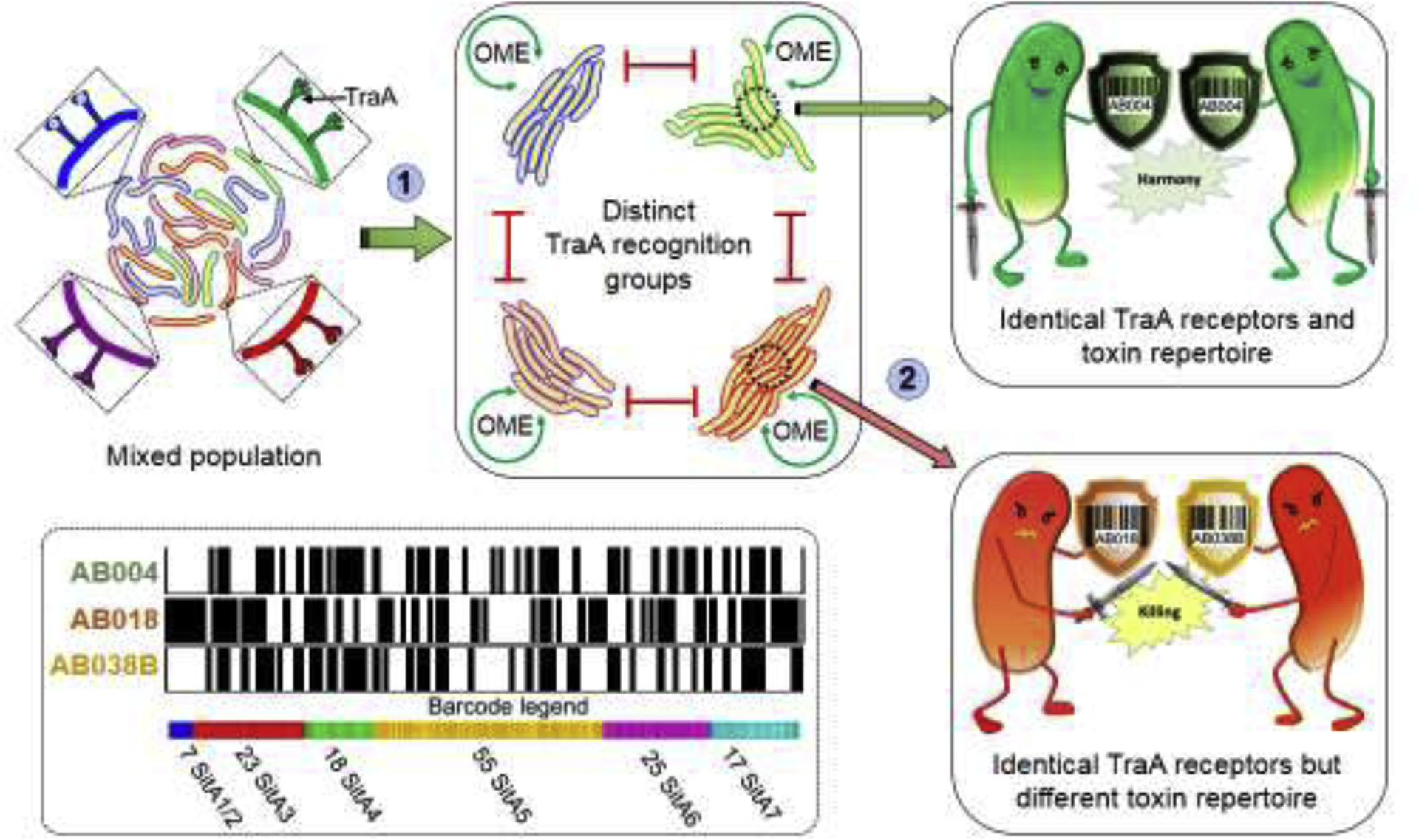
OME of SitA toxins leads to distinct kin groups. Cartoon illustrates two levels of discrimination. (1) Mixed population of myxobacteria discriminate kin from foe by homotypic matching between TraA receptors. Only cells with compatible traA alleles conduct OME. (2) Compatible TraA cells that are clonal contain identical sets of sitAI toxin-immunity loci (barcodes) survive OME and ensures a homogenous population (top). In contrast, cells with compatible TraA receptors that are not clonal contain different toxin-immunity barcodes and annihilate one another (bottom). Bottom left, based on genomic analysis of six wild strains with compatible traA alleles a total of 145 unique sitA loci were identified [30]. Barcodes for three strains shown; black lines indicate the presence of a particular toxin while white lines absence thereof.
