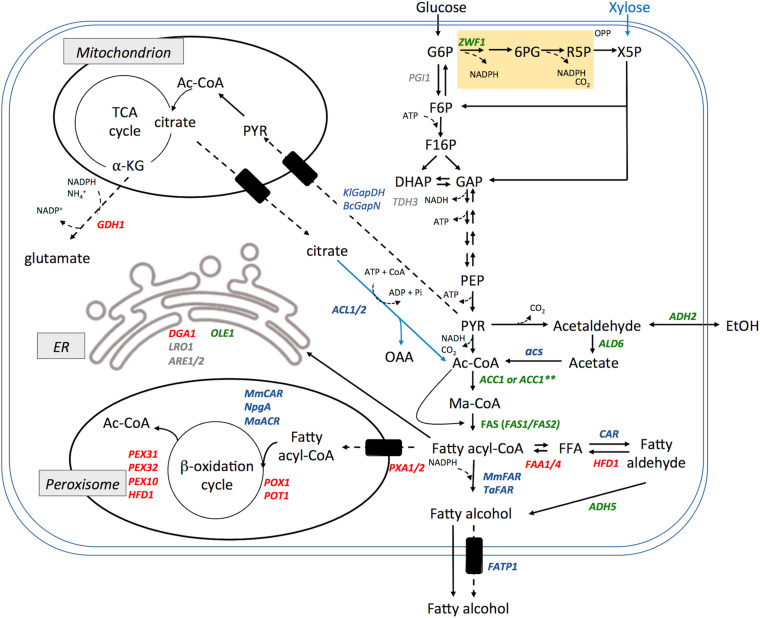
FIGURE 4.
Summary of metabolic engineering performed to increase fatty alcohol production in S. cerevisiae. Heterologous genes expressed (shown in blue), native genes overexpressed (shown in green), native genes deleted or disrupted (shown in red). Other endogenous genes that been manipulated without improving fatty alcohol production are shown in gray. Cofactors are noted in particularly salient positions but are omitted from others for clarity. Note: these enzyme combinations have not necessarily been combined into a single production strain. Abbreviation and gene names used are: G6P, glucose-6-phosphate; 6PG, 6-phosphogluconate; R5P, ribulose-5-phosphate; X5P, Xylulose-5-phosphate; OPP, oxidative pentose phosphate pathway; F6P, fructose-6-phosphate; F16P, fructose-1,6-bisphosphate; DHAP, dihydroxyacetone phosphate; GAP, glyceraldehyde-3-phosphate; PEP, phosphoenolpyruvate; PYR, pyruvate; Ac-CoA, acetyl Co-enzyme A; Ma-CoA, Malonyl CoA; OAA, oxaloacetate; αKG, α-ketoglutarate; FFA, free fatty acid; FAS, fatty acid synthase; ER, endoplasmic reticulum; ZWF1, Glucose-6-phosphate dehydrogenase; PGI, phosphoglucose isomerase; TDH3, Glyceraldehyde-3-phosphate dehydrogenase; KlGapDH, glyceraldehyde-3-phosphate dehydrogenase from Kluyveromyces lactis; BcGapN, non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase from Bacillus cereus; ADH2, alcohol dehydrogenase 2; ALD6, aldehyde dehydrogenase; acs, acetyl CoA synthetase; ACC1, acetyl-CoA carboxylase; ACC1∗∗, A hyperactive ACC1 mutant; ACL1/2, ATP-citrate lyase; GDH1, glutamate dehydrogenase; DGA1, diacylglycerol acyltransferase; LRO1, acyltransferase; ARE1/2, Acyl-CoA:sterol acyltransferase; OLE1, Δ9 fatty acid desaturase; HFD1, fatty aldehyde dehydrogenase; FAA1/4, Long chain fatty acyl-CoA synthetase; ADH5, alcohol dehydrogenase isozyme 5; PXA1/2, peroxisomal ABC-transporter; POX1, fatty acyl-CoA oxidase; POT1, Peroxisomal Oxoacyl Thiolase; PEX10/31/32, peroxisomal membrane proteins; FATP1, human free fatty acid transporter; MmCAR, Mycobacterium marinum carboxylic acid reductase; NpgA, activator protein from Aspergillus nidulans; MaFAR, Marinobacter aquaeolei VT8 acyl-CoA reductase; TaFAR1, Tyto alba fatty acyl-CoA reductase 1; MmFAR1, Mus musculus fatty acyl-CoA reductase 1.