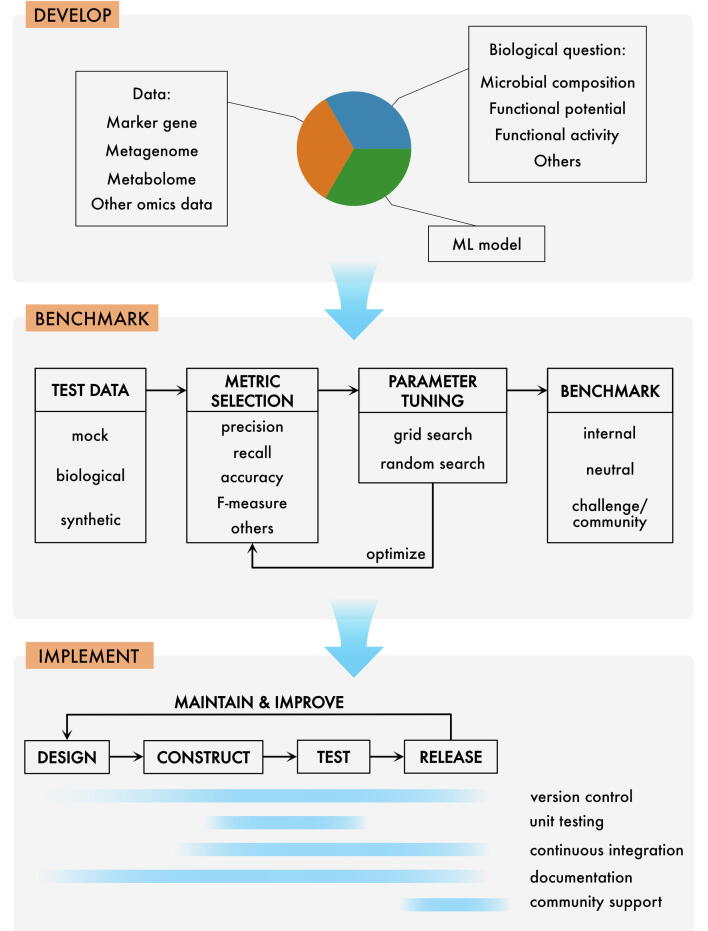
Fig. 1.
An overview of a microbiomics method development workflow. Typically, a method is developed to address one or more biological questions, e.g., concerning microbial composition (genetic or taxonomic), dynamics, or functional activity. Depending on the question, various data types can be used to feed a machine learning model or statistical test(s). Once developed, the method should be subject to a suite of benchmarks to assess its performance. A range of choices should be made here, as to what data to use, which performance metric to apply and what kind of benchmark to employ. Finally, to optimize accessibility for the research community, the method should be implemented into a software package/plugin, applying best practices for software development, including version control, testing and continuous integration, documentation and, finally, community support. Naturally, the three steps presented here may overlap in the development cycle, e.g. some part of benchmarking may be started already in the development phase. Software implementation also often starts early, with the first version of the working code. Generally, however, the transition from “develop” through “benchmark” to “implement” becomes natural as the project progresses.