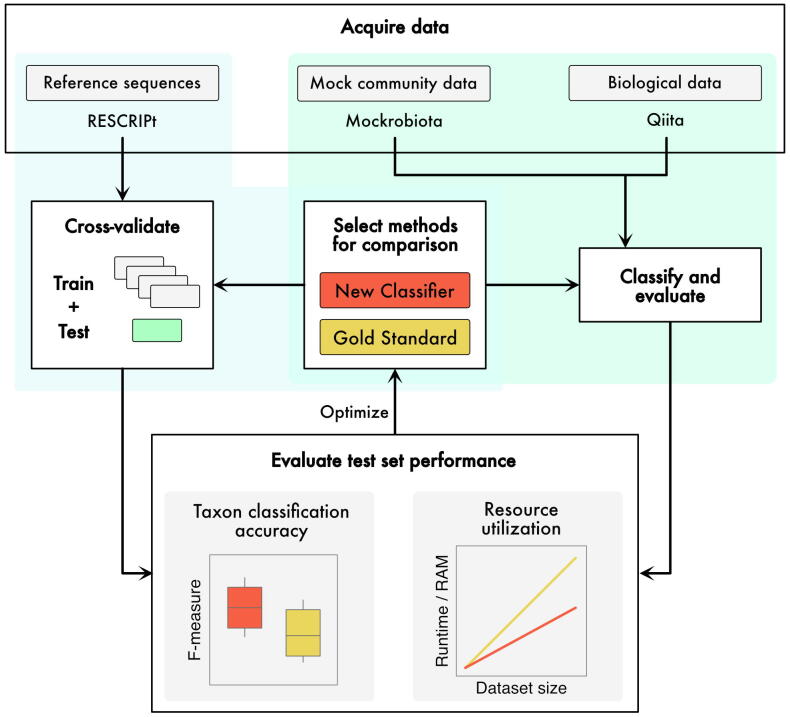
Fig. 3.
An example of a benchmarking workflow for development of a new taxonomic classification method. Test data can be retrieved from multiple sources to obtain: (1) reference sequences for cross validation or simulation (e.g., using RESCRIPt [144]); (2) mock community data and known compositions (e.g., from mockrobiota [123]); and (3) biological data, e.g., microbiome sequence data from Qiita [142]. Data can either be classified directly to evaluate results (e.g., for mock community data, for which the true composition is known), or split into k-folds for cross-validation where at each iteration (k-1) folds are used for model training (represented by grey boxes) and the last fold (green box) is used to evaluate model performance. In the case of taxonomic classification, classification accuracy can be scored using metrics like F-measure. Resource utilization is also recorded and compared to the “Gold Standard” method of choice. If either of the metrics is unsatisfactory, the model can be optimized (e.g., via a grid search of parameter settings) and the process is repeated. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)