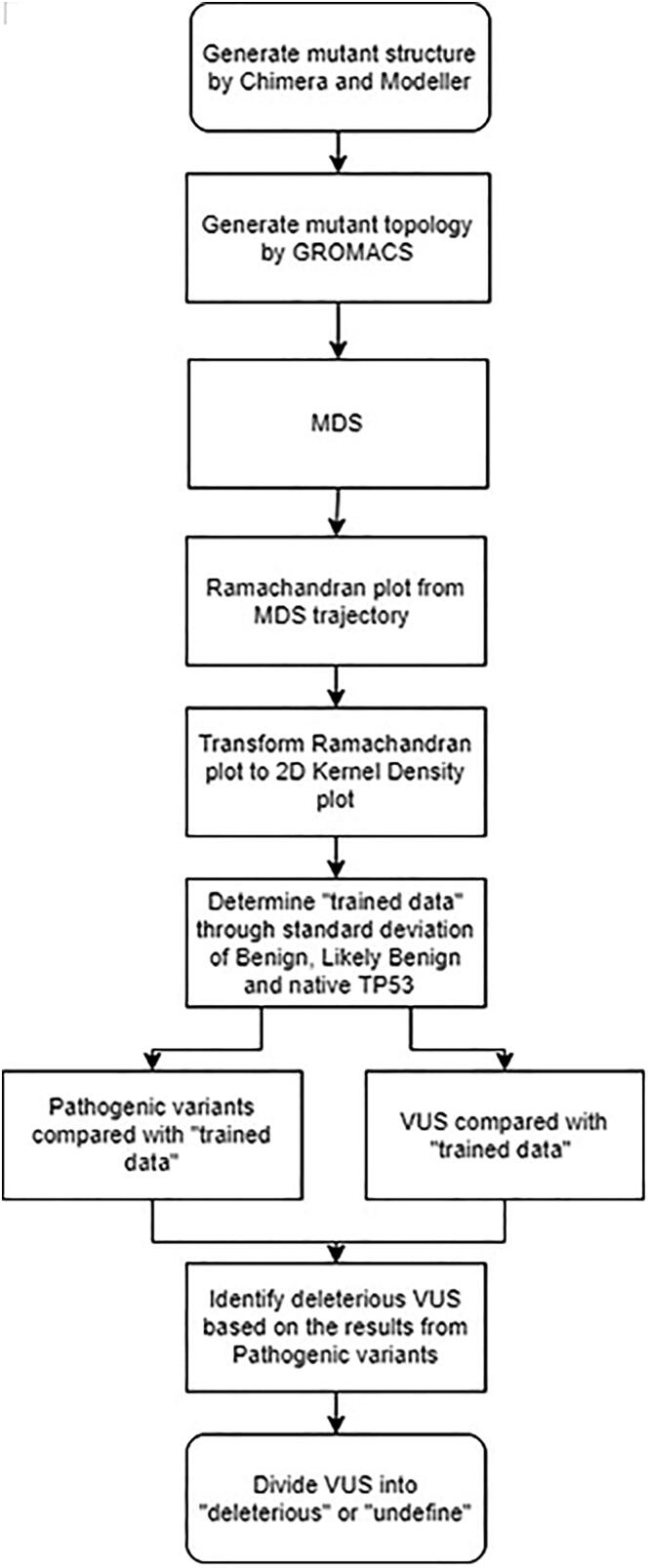
Fig. 1.
Scheme of the study. Starting with the structures of native and mutant protein, each structure was submitted to GROMACS and simulated by MDS. The trajectories from MDS were utilized to create Ramachandran Plot and transformed into density plots by 2D Kernel Density Method. The average deviation of Benign, Likely Benign and native P53 was used as a trained data and compared with Pathogenic variants and VUS. Pathogenic variants were used to create criteria to classify VUS into deleterious and undefined groups.