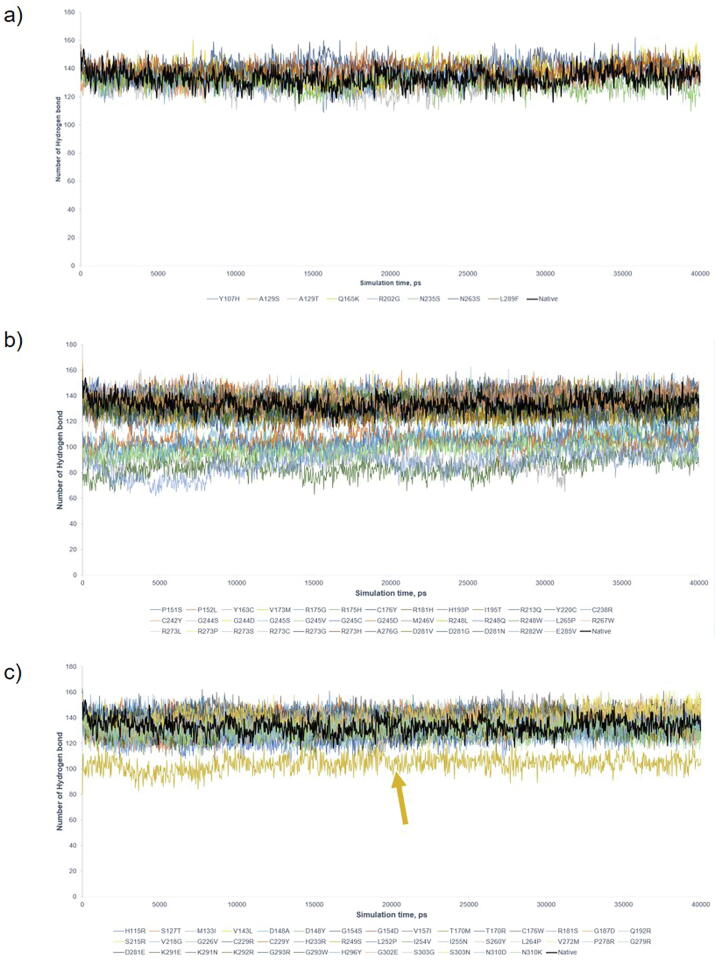
Supplementary Fig. S2.
Hydrogen bond (H bond) graph by 40 ns simulation. The plots were against native P53 (Black). a) 8 Benign/Likely Benign variants; b) 38 Pathogenic variants; and c) 42 VUS. The average H bond was 133 for native P53, 135 ± 4.37 for Benign/Likely Benign, and 135 ± 3.65 for VUS. Pathogenic mutants were separated into 2 regions, the higher and the lower region (Y163C, R175H, Y220C, G245D, G245S, R248Q, R273C, R282W) with 134 ± 4.66 and 98 ± 8.38 H bond, respectively. R249S (Gold) in VUS is identified to have deleterious structure and other VUS showed no significant deviation. The figure showed the H bond alone can only detect the variants with significant deleterious effects on protein structure.