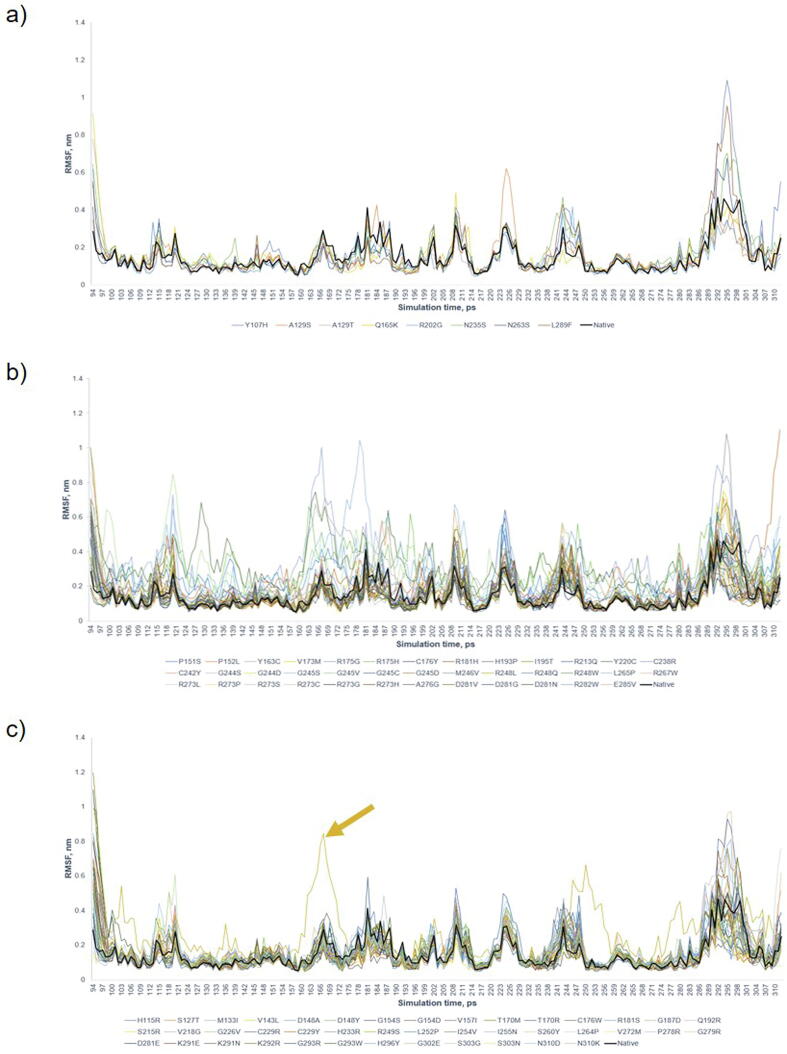
Supplementary Fig. S4.
Root mean square fluctuation (RMSF) graph by 40 ns simulation. a) 8 Benign/Likely Benign variants; b) 38 Pathogenic variants; and c) 42 VUS. All were plotted against native P53 (Black). The Benign/Likely Benign and VUS variants show consistence RMSF in comparison to the native, whereas the region of large difference was between residue 286 – 307. RMSF for Pathogenic mutants has greater fluctuation in comparison to native P53. Only R249S was identified as deleterious and but not for other VUS, indicating that RMSF can only identify the variants with significant deleterious effects on protein structure.