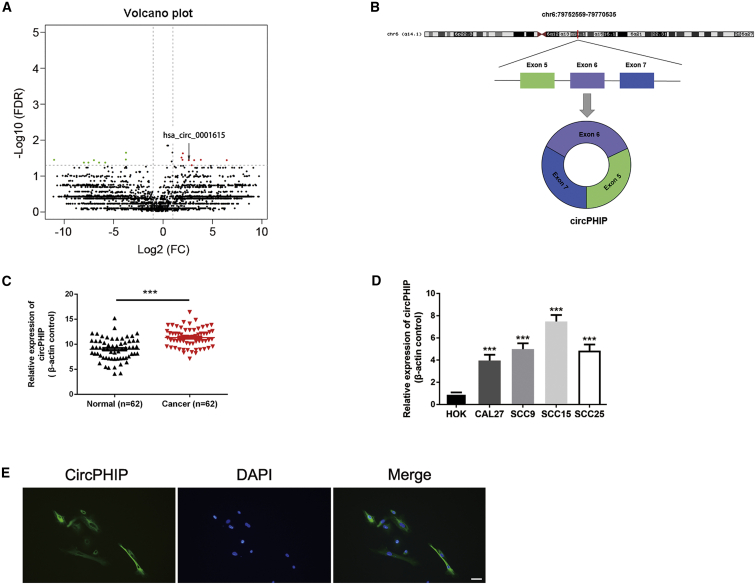
Figure 1.
Identification and validation of differential circPHIP expression in OSCC tissues and cells
(A) Volcano plot of differentially expressed circRNAs. The green dots are downregulated circRNAs, whereas the red dots are upregulated circRNAs in OSCC. circPHIP is labeled as hsa_circ_0001615. (B) Schematic showing the circularization of PHIP exons 5–7 forming circPHIP. (C) circPHIP expression was detected via qRT-PCR in 62 pairs of OSCC and adjacent normal tissues. Student’s t test, ∗∗∗p < 0.001. (D) circPHIP expression was detected via qRT-PCR in various human OSCC cell lines (CAL27, SCC15, SCC9, and SCC25) and healthy oral keratinocytes (HOKs). (E) FISH analysis of circPHIP localization. Scale bar, 50 μm.