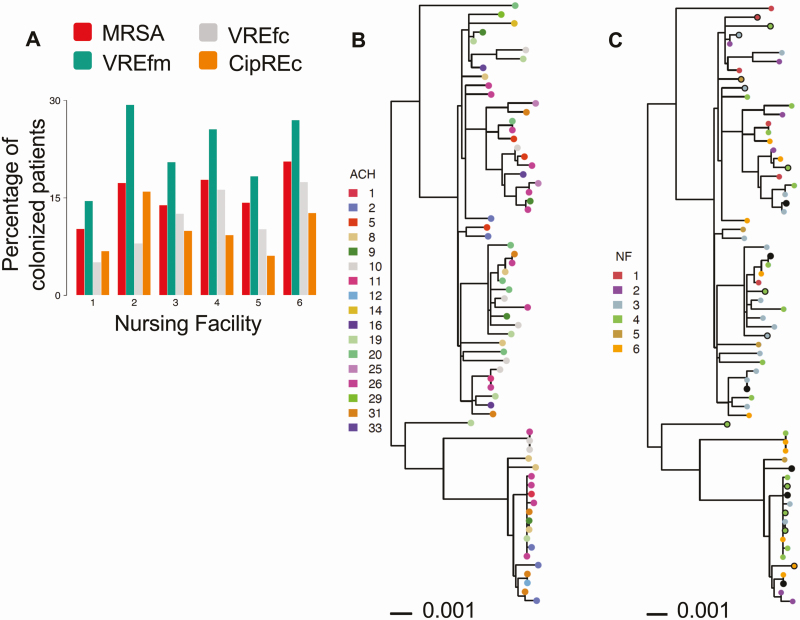
Figure 1.
High prevalence of antibiotic-resistant organisms (AROs) in regional nursing facilities due to endemic spread of epidemic lineages. A, Percentage of patients colonized with prevalent AROs within major healthcare lineages (methicillin-resistant Staphylococcus aureus [MRSA]: sequence type [ST] 5 and ST5-like; vancomycin-resistant Enterococcus faecalis [VREfc]: ST6; ciprofloxacin-resistant Escherichia coli [CipREc]: ST43/131 and ST43/131-like, and clade A vancomycin-resistant Enterococcus faecium [VREfm] isolates) at the time of enrollment or during nursing facility stay, by nursing facility. Each color corresponds to the colonization prevalence of a specific ARO. B and C, Phylogenetic tree of VREfc isolates labeled by patient’s most recent acute care hospital (ACH) exposure (B) and patient’s nursing facility (NF) residence (C) at the time of isolate detection. Isolates collected at the time of admission are shown as solid circles, and those collected during follow-up visits are shown as filled circles. Follow-up isolates pruned from analysis due to close genetic distance with an admission or earlier isolate within the same NF are shown as solid black circles. Sequence type of each isolate is shown on the right. The tree was inferred from maximum likelihood (RAxML) analysis with midpoint rooting. Scale bar represents substitutions per nucleotide site.