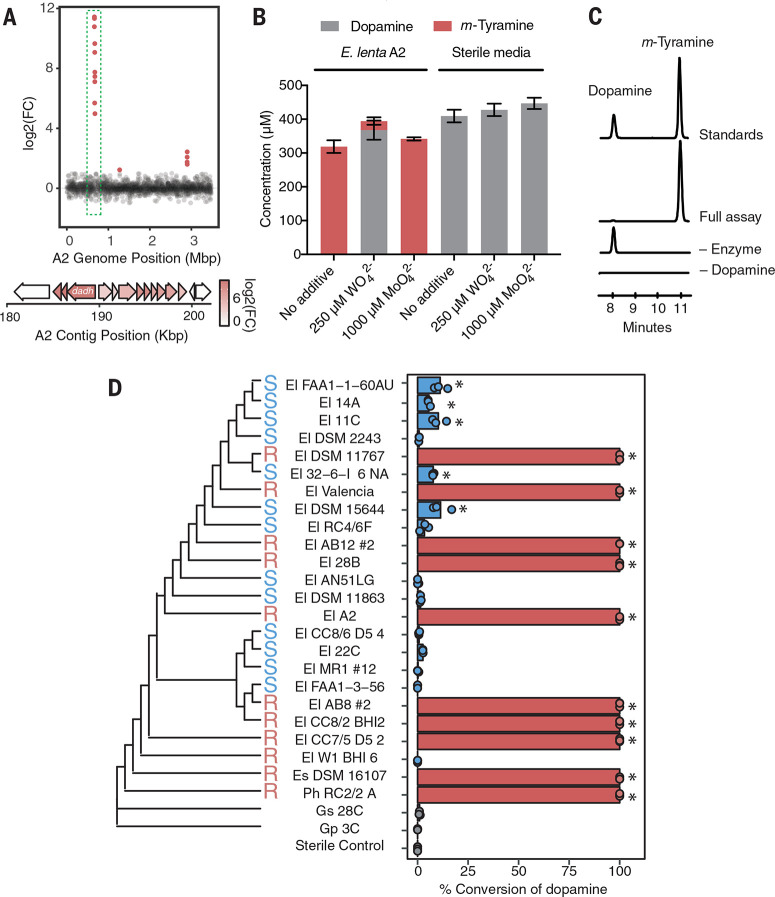
Fig. 2.
E. lenta dehydroxylates dopamine using a molybdenum-dependent enzyme. (A) RNAsequencing identifies a putative molybdenum (moco)–dependent dopamine dehydroxylase (Dadh) in E. lenta A2. Differentially expressed candidate genes (false discovery rate < 0.1 and fold change > j2j) are plotted as a function of genome position, revealing three discrete loci of differentially expressed genes. (Inset) Analysis of the largest cluster of differentially expressed genes at 0.665 Mbp in the scaffolded assembly (190 kg base pairs in the reference contig) revealed that a putative dadh was up-regulated by 2568-fold in response to dopamine. (B) Tungstate treatment inhibits dehydroxylation of dopamine by E. lenta A2. Cultures were grown anaerobically with tungstate (WO42–) or molybdate (MoO42–) for 48 hours with 0.5 mM dopamine. Bar graphs represent the mean ± SEM of three biological replicates. (C) In vitro activity of Dadh-containing fractions purified from E. lenta A2. Extracted LC-MS/MS ion chromatograms for simultaneous detection of dopamine and m-tyramine after 12 hours of anaerobic incubation of enzyme preparation with 500 mM dopamine and artificial electron donors at room temperature. Peak heights show the relative intensity of each mass, and all chromatograms are shown on the same scale. (D) A single amino acid variant predicts dopamine metabolism by E. lenta and related strains (P = 0.013 Fisher’s exact test) and does not correlate with phylogeny. Strains were cultured anaerobically with 500 mM dopamine for 48 hours (El, E. lenta; Es, Eggerthella sinensis; Gs, Gordonibacter sp.; and Gp, Gordonibacter pamelaeae; Ph, Paraeggerthella hongkongensis). High (100% conversion) and low (<11% conversion) metabolizers are denoted in red and blue. For each strain, data points represent biological replicates (P *< 0.05 analysis of variance with Dunnett’s test versus sterile controls).