Figure 3. Systematic Prediction of Targets for the HRD1 Branch of ERAD.
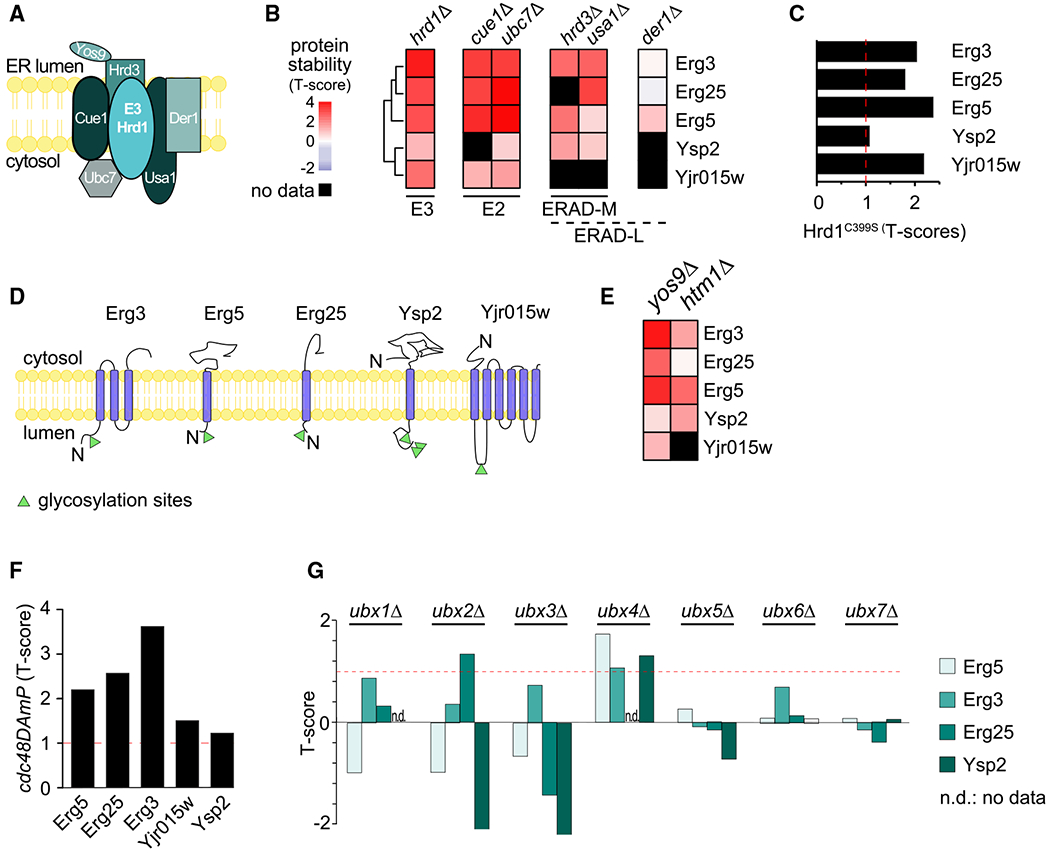
(A) Schematic of the Hrd1 branch of ERAD in yeast.
(B) Hierarchically clustered heatmap of the robust T scores of Hrd1 targets in response to deletion of the member of the Hrd1 complex.
(C) Stability of Hrd1 targets in cells expressing Hrd1C399S, a catalytically dead allele of HRD1.
(D) Localization and topology of bona fide substrates of the Hrd1 complex. Glycosylation sites are indicated as green triangles.
(E) Heatmap clustered and colored as in (B) of the robust T scores of Hrd1 targets in response to the deletion of YOS9 and HTM1.
(F) Stability of Hrd1 complex substrates in cells expressing cdc48DamP, a hypomorphic allele of CDC48.
(G) Stability of Erg5, Erg3, Erg25, and Ysp2 in cells deleted for individual UBX domain-containing substrate adaptors of Cdc48.
