Figure 1. Mtb enoyl-CoA hydratase EchA19 is a homotrimer with conserved active site residues and CoA binding residues and large binding pockets to accommodate cholesterol metabolite substrates.
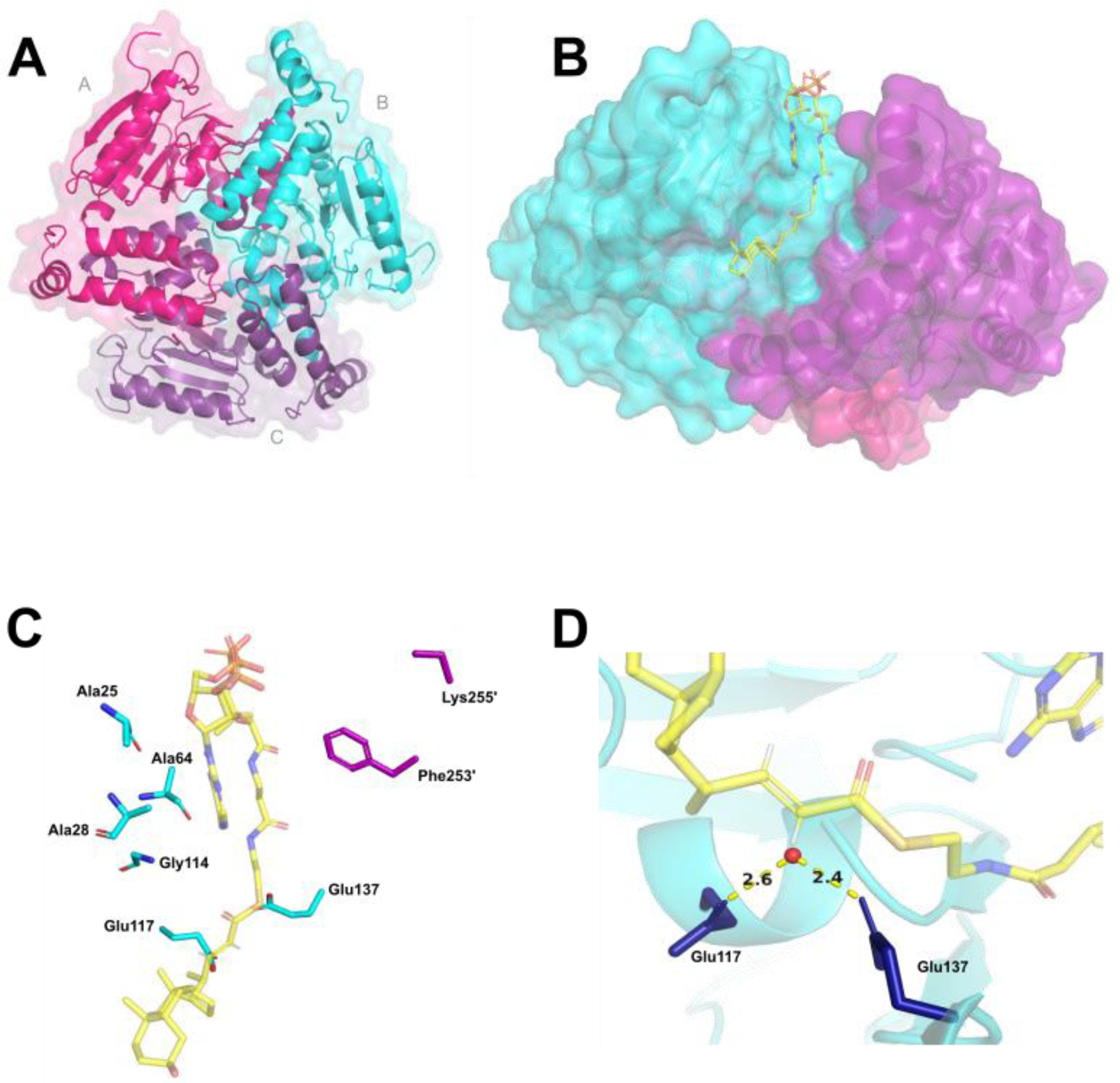
A) KstR1-regulated EchA19 is a homotrimer (monomer A, magenta; monomer B, cyan; monomer C, purple) with three substrate binding pockets formed by the monomer–monomer interfaces. Each monomer possesses the canonical αββ protein fold of the crotonase-like family. B) 3-OCDO-CoA is modeled into an active site of Mtb EchA19. 3-OCDO-CoA adopts an elongated conformation within the active site (monomers B and C are shown). C) Conserved active site residues (Glu117 and Glu137) and substrate interacting residues in the substrate binding pocket are Ala25, Ala28, Ala64, Leu68, Lys69, and Gly114 (which belong to monomer B) and Lys255’ and Phe253’ (which belong to monomer C). The Leu68 and Lys69 residues are located in a disordered region of the Mtb EchA19 crystal structure (Gly65–Arg89). The shortened side chain of Lys255’ is the result of incomplete electron density for this residue in the protein structure. D) Catalytic residues Glu117 and Glu137 coordinate a water molecule (depicted as a red circle), positioning it for addition to the alkene of 3-OCDO-CoA.
