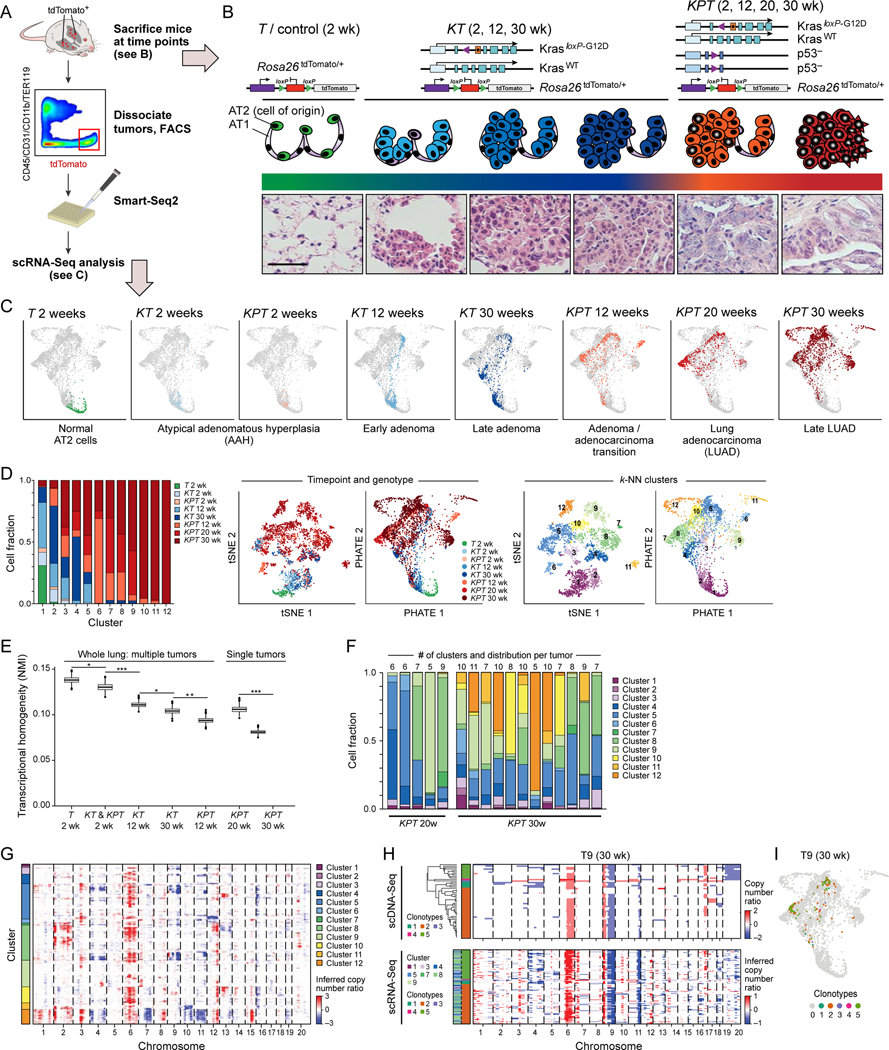
Figure 1. Increased transcriptional heterogeneity in mouse lung adenocarcinoma (LUAD) evolution is reproducible across individual tumors and mice, but cannot fully be explained by gene copy number variation (CNV).
(A) Experimental pipeline. (B) Tumor evolution in a LUAD GEMM. Top: genetic constructs of three mouse models profiled by scRNA-Seq at different time points. Middle and bottom: schematic (middle) and hematoxylin & eosin staining of tissue sections (bottom) at different phases of tumor progression. AT1: normal alveolar type 1 (AT1) cells; AT2: normal alveolar type 2 (AT2) cells, AAH: atypical adenomatous hyperplasia. Scale bar: 100 μm. (C) PHATE map embedding (STAR Methods) of scRNA-Seq profiles (dots) collected from the models and time points in (B) (labels, top). Colored dots: Cells collected from the indicated sample; grey dots: all other cells. (D) Increased diversity of cell clusters with progression. Left: The fraction of cells (y axis) in each cluster (x axis) that are derived from each sample type (genotype and time point; colored as in (C)). Middle and Right: matched t-stochastic neighbor embedding (tSNE, left plot, STAR Methods) and PHATE map embedding (right plot, as in (C)) colored by either sample type (middle pair) or cluster number (STAR Methods) (right pair). (E) Reduced transcriptional homogeneity within time point with progression. Transcriptional heterogeneity is inversely proportional to the Normalized Mutual Information (NMI, y axis) between cells within in each sample type (genotype/time point combination, x axis), for either whole lung samples or microdissected single tumors. Box plots: upper, median, and lower quartile of 1,000 bootstrap samples, of 50 cells each, from the indicated time point; whiskers: 1.5 interquartile range. * p < 0.05, ** p < 0.01, *** p < 0.001 (STAR Methods). (F) Fraction of cells (y axis) in sample (x axis) that are members of each cluster (color code, as in D, right). The number of clusters observed in each individually plucked tumor is indicated at the top of the bars. (G) CNVs (red: amplifications, blue: deletions) across the chromosomes (columns) inferred from the scRNA-Seq of each cell (rows) from 12 KP tumors at the 30-week time point (STAR Methods). Color: the cluster membership of each cell. (H) Congruence between CNV profiles inferred from scDNA-Seq and scRNA-Seq. CNVs shown as in (G) for single cells (rows) of one individually microdissected KPT tumor at 30 weeks profiled by scDNA-Seq (top-left) or scRNA-Seq (bottom-left). Left color bar: Predominant clonotypes identified from scDNA-Seq (top-left) and assigned to scRNA-Seq cells (bottom-left). Far left color bar in scRNA-Seq panels: cell cluster membership as in (G). (I) A single clonotype matches multiple transcriptional states. PHATE map as in Figure 1D, colored by clonotype. See also related Figure S1.