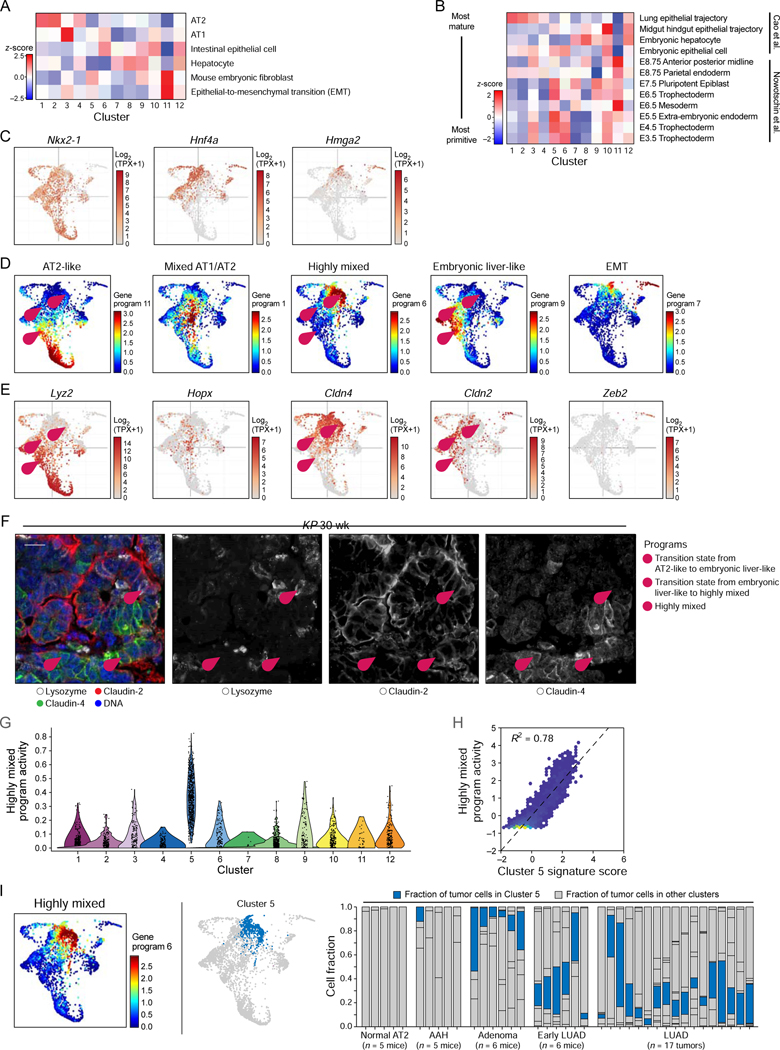
Figure 2. Loss of lung lineage fidelity in LUAD progression and emergence of a highly mixed identity program.
(A, B) Signature score (color bar, STAR Methods) of either adult [(Han et al., 2018; Zhang et al., 2019); (A), z-score)] or embryonic [(Cao et al., 2019; Nowotschin et al., 2019); (B), z-score)] mouse cell signatures in the cells of each cluster (columns). In (B), signatures (rows) are ordered from most differentiated (top) to most primitive (bottom) cells. (C) PHATE maps (as in Figure 1D), with cells (dots) colored by expression (Log2(TPX+1), color bar) of Nkx2–1, Hnf4a, and Hmga2. (D, E) Five key gene programs highlight alternative cell type programs, two key transition states and an EMT-like state. PHATE map (as in Figure 1D), with cells (dots) colored either by the activity of each program (D, NMF loading, color bar, see Figure S2C for additional programs, STAR Methods) or by the expression level (E, Log2(TPX+1), color bar) of a selected marker from the corresponding program. (F) Immunofluorescence for Lysozyme (AT2-like program), Claudin-2 (hepatocyte-like program), and Claudin-4 (highly mixed program). Pink numbered arrowheads indicate cell states or transitions in (D-F): 1 - AT2-like (lysozyme) to Embryonic liver-like (Claudin-2) transition; 2 – Embryonic liver-like (Claudin-2) to Highly mixed (Claudin-4) transition; 3 - Highly mixed program (Claudin-4). Scale bar: 20 μm. (G) Cells from cluster 5 show significantly elevated activity of the Highly mixed NMF program (t-test, p < 1×10 −16). (H) Cell scores for Highly mixed program (y axis) and a cluster 5 signature (x axis). Pearson R2 = 0.78. Lighter dot color indicates higher cell density. (I) PHATE map embedding as in Figure 1D, with cells (dots) colored by score of the highly mixed program (left) or by cluster 5 membership (blue, middle). Right: Proportion of cells (y axis) from cluster 5 (blue) in each sample (mouse or tumor; x axis), ordered by tumor progression. See also related Figure S2 and Table S2.