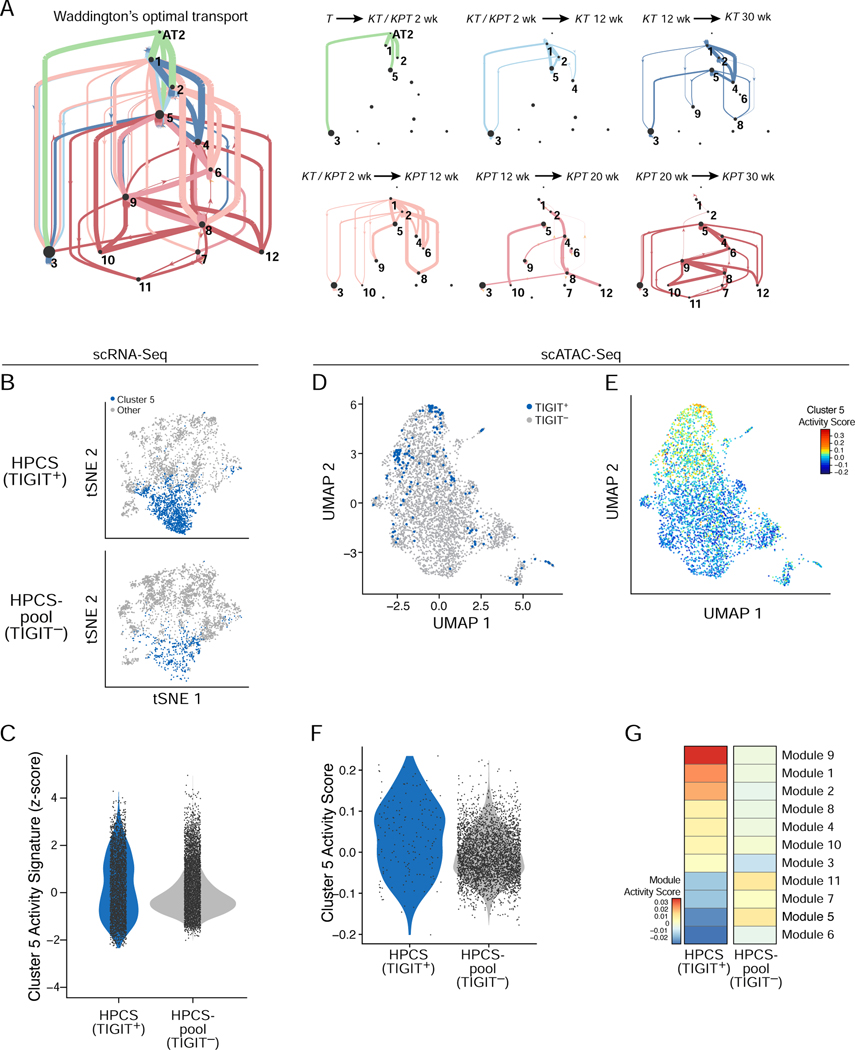
Figure 3. Identification of a highly plastic cell state with a distinct chromatin accessibility profile.
(A) Probability of cell state transitions as predicted by an optimal transport model. Two cell clusters (nodes, proportional to ‘pagerank’ score – proportion of time spent at node on a random walk) A and B are connected by a directed edge from A to B, if the cells in cluster A at time point t (color code, as in Figure 1B,C) are predicted by the optimal transport model to be ancestors of cells in cluster B at the next time point in that model. Edge thickness is proportional to the probability of the transition predicted by the model (low probability edges < 0.1, are excluded for graphical clarity). Right: Sub-graphs showing only edges between clusters for selected time couplings (labels, top) are on the right. Line width is proportional to the probability of transition ranges from <0.01 for the thinnest line to 0.65 for the thickest line. Dot size is proportional to the pagerank importance of each node, i.e. the amount of “time” spent in a random walk on the graph in any given node. (B) tSNE of cell profiles from primary tumor cells sorted as TIGIT+ (top) and TIGIT− (bottom) sampled to the same cell numbers, colored by membership in cluster 5 (blue). Cells sorted from n = 12 mice. (C) Distribution of cluster 5/HPCS signature score (y axis) in TIGIT+ and TIGIT− cells (p = 3.08 × 10−25; Mann-Whitney U test). (D) UMAP embedding of scATAC-Seq profiles from 164 TIGIT+ (blue) and 3,787 TIGIT− (grey) cells from dissociated primary tumors of n = 5 mice (E) UMAP as in (D) but with cells colored by cluster 5/HPCS gene activity signature scores. (F) Distribution of cluster 5/HPCS gene activity signature score (y axis) in scATAC-Seq profiles of TIGIT+ and TIGIT− cells from n = 5 mice (p = 1.8×10−6, Wilcoxon rank-sum test). (G) Activity scores (color bar) of chromatin state modules (rows, from LaFave et al.) in TIGIT+ and TIGIT− sorted cells (columns) from n = 5 mice. See also related Figure S3 and Table S3.