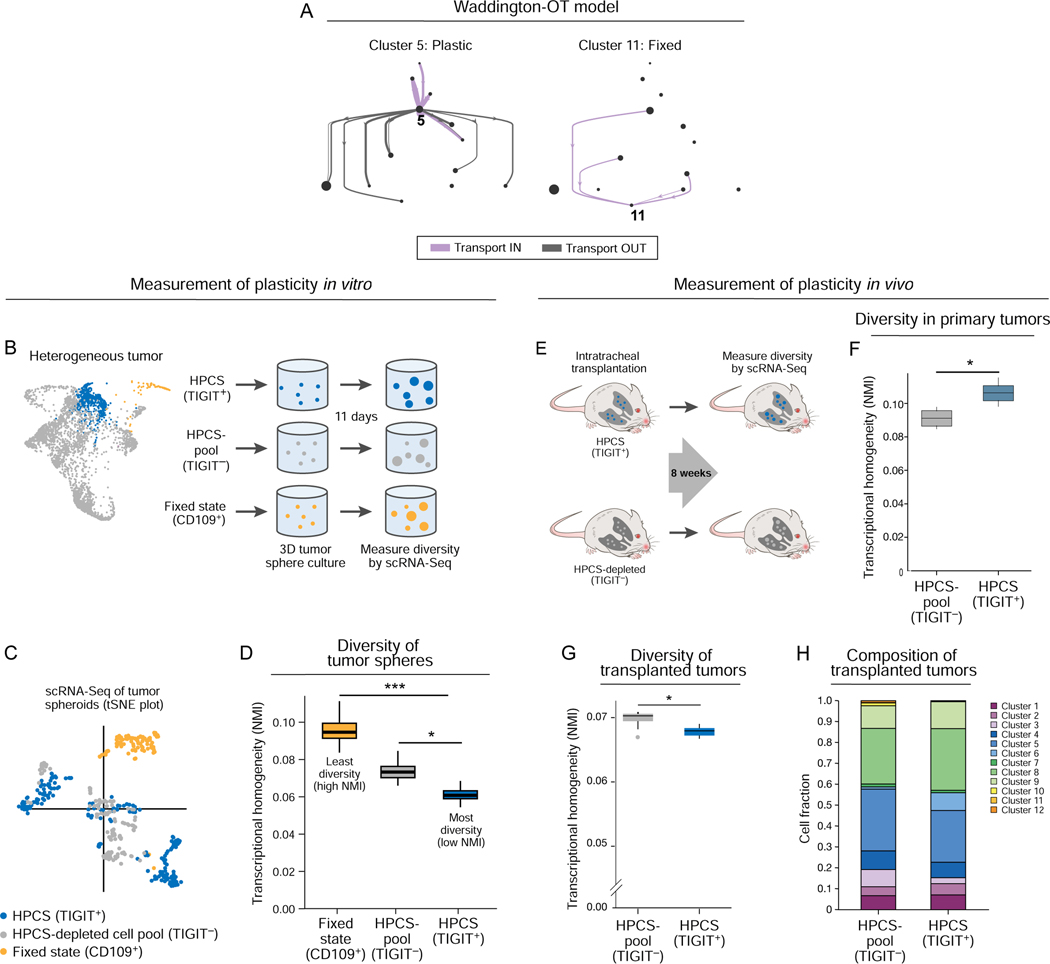
Figure 4. Prospectively isolated HPCS cells display high differentiation potential in vitro and in vivo.
(A) Prediction of plastic and static cell states by the optimal transport model. Graph as in Figure 3A, but showing all transitions (aggregate across all time points) to and from cluster 5 (left) or 11 (right) cells. (B) Experimental design. TIGIT+ HPCS/cluster 5 cells (blue), CD109+(cluster 11) cells (gold), and all non-HPCS TIGIT− cells (grey) were sorted from 17–22 week old LUAD tumors, and grown as tumor spheres for 11 days, followed by scRNA-Seq. (C) tSNE of scRNA-Seq profiles of cells from tumor spheres arising from TIGIT+ (blue), CD109+ (gold) and TIGIT− (grey) KP or KPT LUAD cells at 11 days after cell plating (n = 7 mice). (D) Transcriptional homogeneity. Normalized Mutual Information (NMI, y axis) between each of the three populations. Box plots: upper, median, lower quartile of 1,000 bootstrap samples, of 50 cells each, from the indicated time point; whiskers: 1.5 interquartile range. * p < 0.05, *** p < 0.001 (STAR Methods). (E) Experimental design. TIGIT+ HPCS/cluster 5 cells (blue) and all non-HPCS TIGIT− cells (grey) were sorted from 18–21 week LUAD tumors, and orthotopically transplanted to lungs of NSG mice. (F) Normalized Mutual Information (NMI, y axis) within TIGIT+ and TIGIT− populations. Box plots: upper, median, lower quartile of 1,000 bootstrap samples, of 100 cells each, from the indicated time point; whiskers: 1.5 interquartile range. * p < 0.05 (n = 2 biological replicates, STAR Methods) (n = 6 mice). (G) NMI (y axis) between each population. Box plots: upper, median, lower quartile of 1,000 bootstrap samples, of 50 cells each, from the indicated time point; whiskers: 1.5 interquartile range. * p < 0.05. (H) Relative proportion of cells from TIGIT+ and TIGIT− transplanted primary tumor cells in each cluster (n = 5 TIGIT− vs 3 TIGIT+ allotransplant mice).