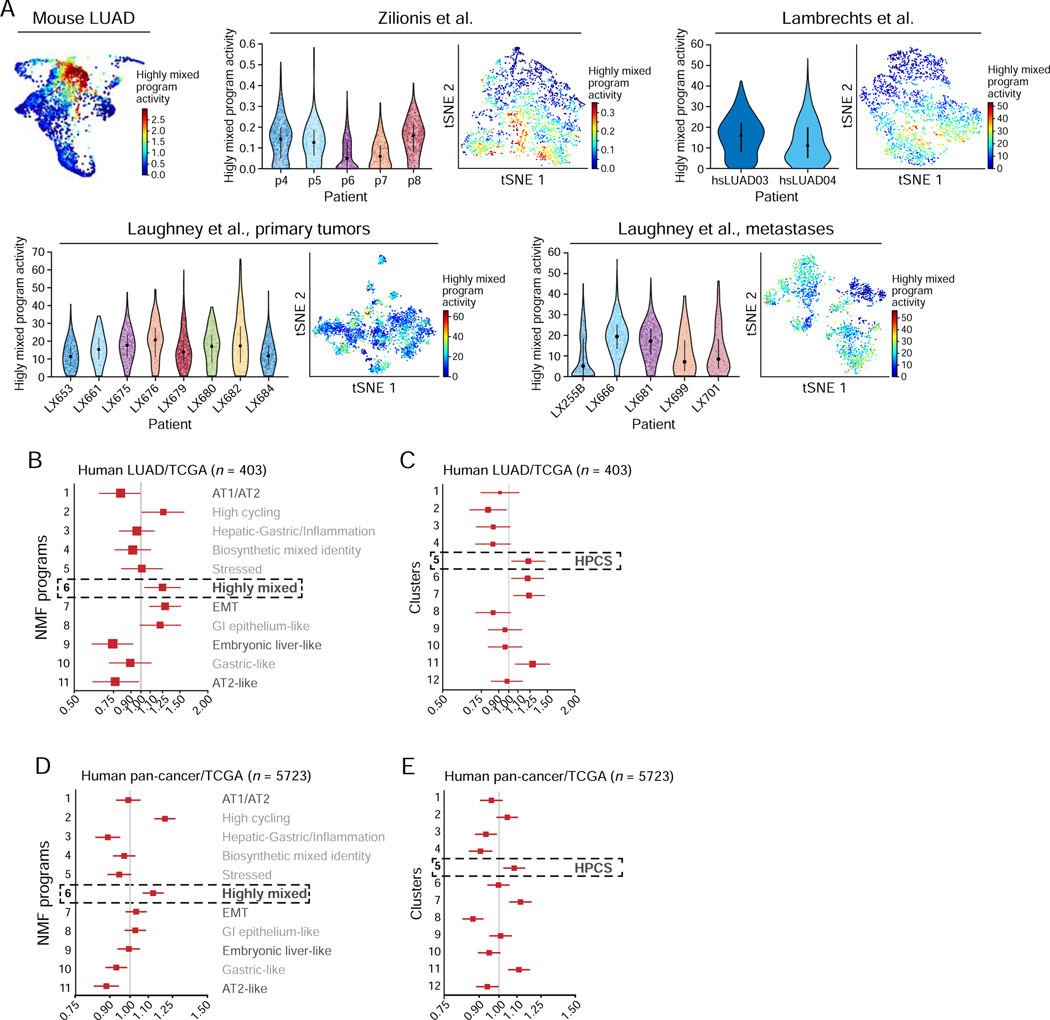
Figure 6. HPCS-like program is expressed in human tumors and associates with poor survival.
(A) The high-plasticity program is expressed in individual malignant cells from human LUAD tumors. Left: PHATE map of the mouse LUAD cells (as in Figure 2D), colored by the program score. Right: For each of three scRNA-Seq studies of cancer cells from human LUAD tumors, shown are the violin plot (left) of the distribution of the Highly mixed/HPCS program scores (y axis) in the cancer cells of each tumor (x axis), and a tSNE of the profiles, with cells (dots) colored by their program scores. (B) Hazard ratio (HR, x axis, mean HR and 95%-confidence interval) for each NMF program (y axis) in LUAD patients in the TCGA as predicted by a Cox proportional hazards model independently fit to each NMF activity term as a continuous variable (n = 403; STAR Methods). (C) Hazard ratio (HR, x axis, mean HR and 95%-confidence interval) for each cluster (y axis) in LUAD patients in the TCGA as predicted by a Cox proportional hazards model independently fit to each cluster activity term as a continuous variable (n = 403; STAR Methods). (D) Hazard ratio (HR, x axis, mean HR and 95%-confidence interval) for each NMF program (y axis) across all tumors with tumor purity information in TCGA (n = 5723) as predicted by a Cox proportional hazards model independently fit to each NMF activity term as a continuous variable (STAR Methods). (E) Hazard ratio (HR, x axis, mean HR and 95%-confidence interval) for each cluster (y axis) in all cancer patients in the TCGA as predicted by a Cox proportional hazards model independently fit to each cluster activity term as a continuous variable (n = 5723; STAR Methods). See also related Figure S5 and Table S5.