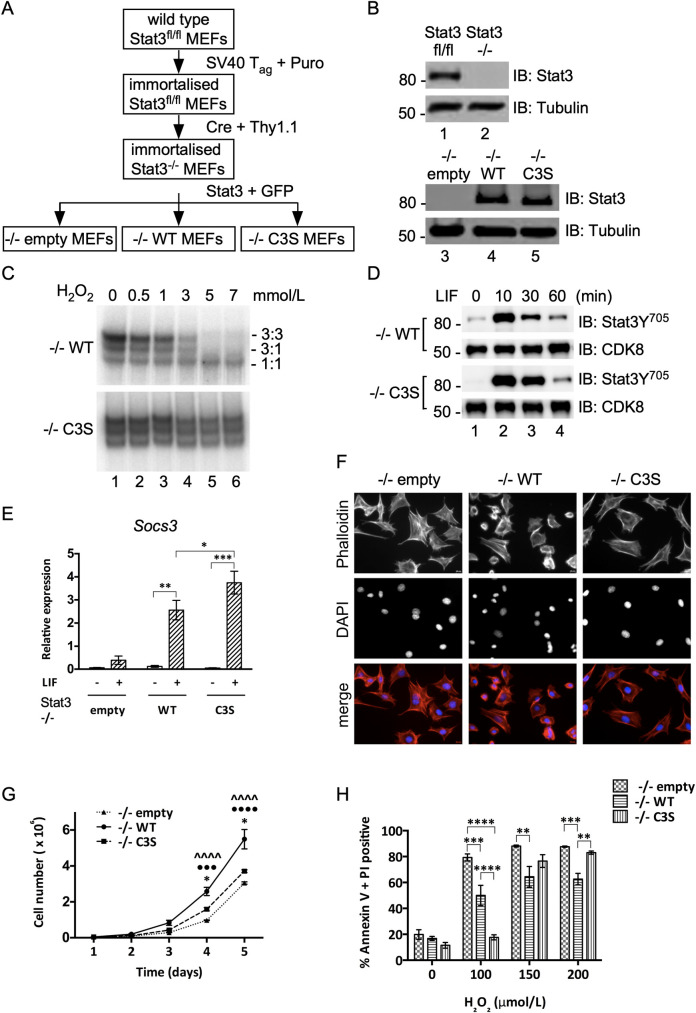
Fig 1. Generation and characterization of Stat3 cell lines.
A) Flow diagram for the production of Stat3 null (-/-empty), WT-Stat3 (-/-WT) and redox-insensitive Stat3-C3S (-/-C3S) expressing MEFs. B) Upper: Immunoblot confirming Stat3 deletion Lower: Immunoblot confirming re-expression of Stat3-WT and Stat3-C3S. C) Nuclear extracts from -/-WT and -/-C3S MEFs were incubated with increasing concentrations of peroxide and binding to a radio-labelled m67/SIE was analysed by EMSA. DNA-bound Stat 3:3, 3:1 and 1:1 homo- and heterodimers are indicated. D) Induction of Stat3 Y705 phosphorylation in both -/-WT and -/-C3S MEFs upon LIF treatment. Serum starved MEFs were treated with 10ng ml-1 LIF and harvested at times indicated. Stat3 Y705 phosphorylation was detected by SDS-PAGE and immunoblotting with an anti-phospho-Y705 antibody. E) Induction of Socs3 expression in -/-WT and -/-C3S MEFs upon LIF treatment. qRT-PCR analysis was performed on Socs3 and Hbs1l mRNA enriched from total RNA from MEFs untreated (-) or treated with 10 ng ml-1 LIF for 30 mins (+) using Taqman assays. Socs3 expression was normalised to Hbs1l expression. Data are expressed as mean ± SEM, n = 3. F) Phalloidin staining to show the arrangement of actin fibres in -/-empty, -/-WT and -/-C3S MEFs. Cells were stained with 165 nM Alexa Fluor 488 phalloidin and imaged at 20 x magnification. G) MEFs were seeded in 6-well plates and grown for up to 120 h under basal conditions. Cells were counted every 24h. H) MEFs were seeded and grown for 16 h under normal conditions. Cells were treated with peroxide at the concentrations indicated and levels of PCD were determined using Muse Annexin V and dead cell assay kit after 6 h, collecting 2000 cells per run. Data are expressed as mean ± SEM, n = 3.