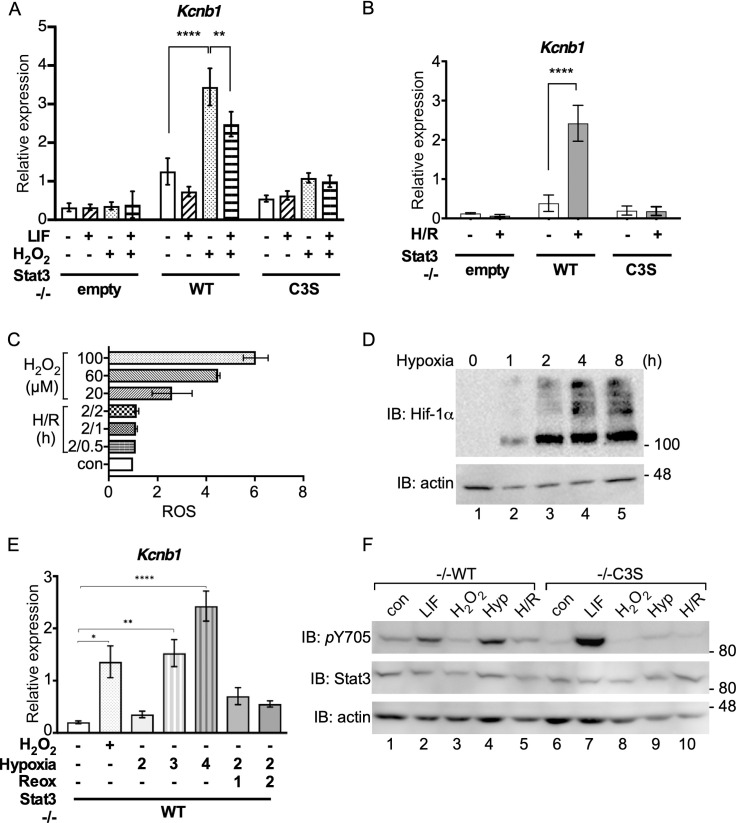
Fig 5. Kcnb1 expression in response to hypoxia/reoxygenation requires Stat3 oxidation.
A) RNA isolated from MEFs untreated (-), treated with 10 ng ml-1 LIF for 30 mins and/or 100 μM H2O2 for 1 h (+) was enriched for mRNA and analysed for Kcnb1 and Hbs1l expression by qRT-PCR. Kcnb1 expression was normalised to Hbs1l. Data are expressed as mean ± SEM, n = 3. B) RNA isolated from MEFs untreated (-) or subjected to 2 h hypoxia (1% O2) and 2 h reoxygenation (H/R) was enriched for mRNA and analysed for Kcnb1 and Hbs1l expression by qRT-PCR. Kcnb1 expression was normalised to Hbs1l. Data are expressed as mean ± SEM, n = 3. C) -/- WT MEFs seeded to 96-well plates (1 x 104 per well) and cultured under basal conditions overnight were stained with 100 μM DCFDA for 30 minutes and left to recover in full medium for 1 h when basal fluorescence was measured. Cells were treated with 0, 20 μM, 60 μM and 100 μM H2O2 or 2 h hypoxia followed by 30 mins, 1 h and 2 h reoxygenation, as indicated, before fluorescence measurement. Values are presented as means ± SEM from 3 independent determinations. D) -/-WT MEFs were exposed to hypoxia (1% O2) for the times indicated. Whole cell extracts were prepared and analysed for stabilisation of Hif-1α by SDS-PAGE and immunoblotting. E) -/- WT MEFs were subjected to normoxia (untreated, -), 100 μM H2O2, hypoxia for 2, 3 and 4 h and hypoxia for 2 h followed by 1 h and 2 h of reoxygenation (treated, +). qRT-PCR analysis was performed using mRNA enriched from total RNA. Expression of Kcnb1 was normalised to expression of Hbs1l. Values are presented as mean ± SEM, n = 3 per group. *P<0,1, **P<0.01, ****P<0.0001. F) Whole cell lysates from -/-WT and -/-C3S MEFs untreated (con), treated with 10 ng ml-1 LIF for 30 mins (LIF), 100 μM H2O2 for 1 h (H2O2), 2 h hypoxia (1% O2; Hyp) or H/R as in (B) were analysed for Stat3 Y705 phosphorylation by SDS-PAGE and immunoblotting.