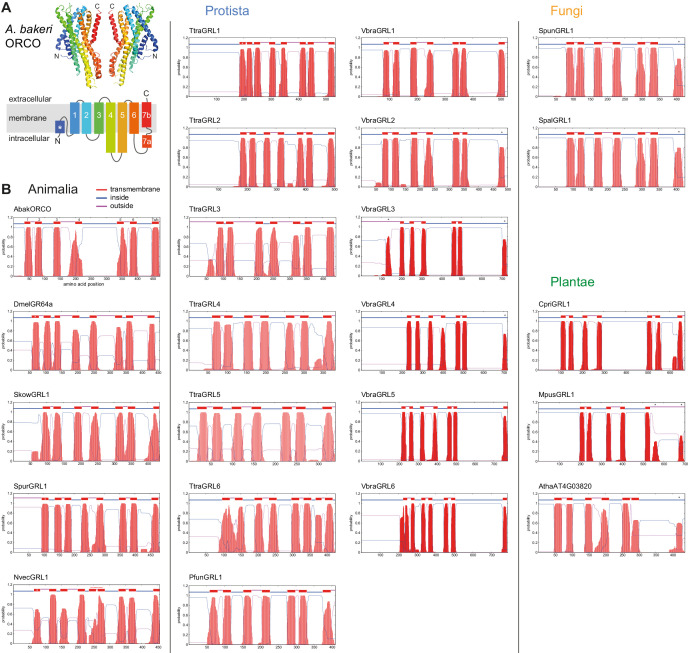
Figure 1. Transmembrane topology predictions of GRLs.
(A) Top: cryo-EM structure of Apocrypta bakeri ORCO (AbakORCO) (PDB 6C70 [Butterwick et al., 2018]); only two subunits of the homotetrameric structure are visualized. Bottom: Schematic of the membrane topology of AbakORCO (adapted from Butterwick et al., 2018), colored as in the cryo-EM structure. The white asterisk marks a helical segment that forms part of a membrane re-entrant loop in the N-terminal region. TM domain seven is divided into a cytoplasmic segment (7a) and a membrane-spanning segment (7b). (B) TM domain and topology predictions of the previously described and newly-recognized GRLs and DUF3537 proteins (Dmel, Drosophila melanogaster; Skow, Saccoglossus kowalevskii; Spur, Strongylocentrotus purpuratus; Nvec, Nematostella vectensis; Atha, Arabidopsis thaliana; see Table 1 for other species abbreviations and sequence accessions). Each plot represents the posterior probabilities of transmembrane helix and inside/outside cellular location along the protein sequence, adapted from the output of TMHMM Server v2 (Krogh et al., 2001). In several sequences an extra transmembrane segment near the N-terminus is predicted (marked by a white asterisk in the N-best prediction above the plot); this may represent the re-entrant loop helical region observed in ORCO, rather than a transmembrane region; in at least one case (SpurGRL1) the designation of this region as a TM domain, leads to an atypical (and presumably incorrect) prediction of an extracellular N-terminus. Conversely, in a subset of proteins individual TM domains are not predicted (notably TM7, black asterisks above the N-best plot), which is likely due to subthreshold predictions for TM domainsin these regions. In NvecGRL1, the long TM4 helix (which projects into the intracellular space in ORCO [Butterwick et al., 2018]) is mis-predicted as two TM domains (dashed red line). Independent membrane topology predictions for unicellular species’ GRLs were obtained using TOPCONs (Supplementary file 2), with largely consistent results.