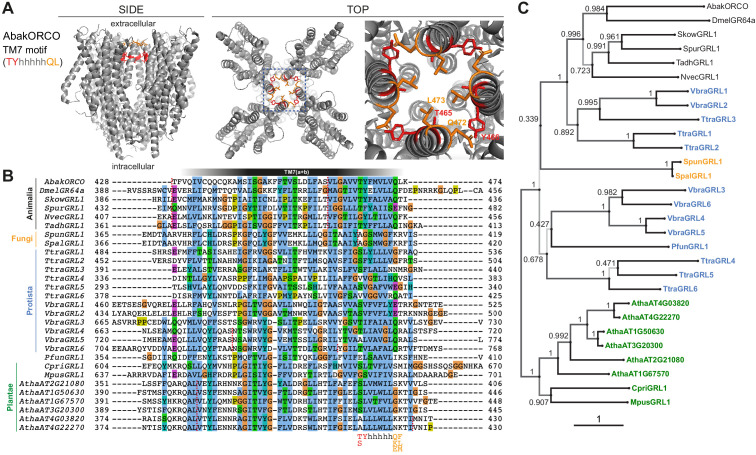
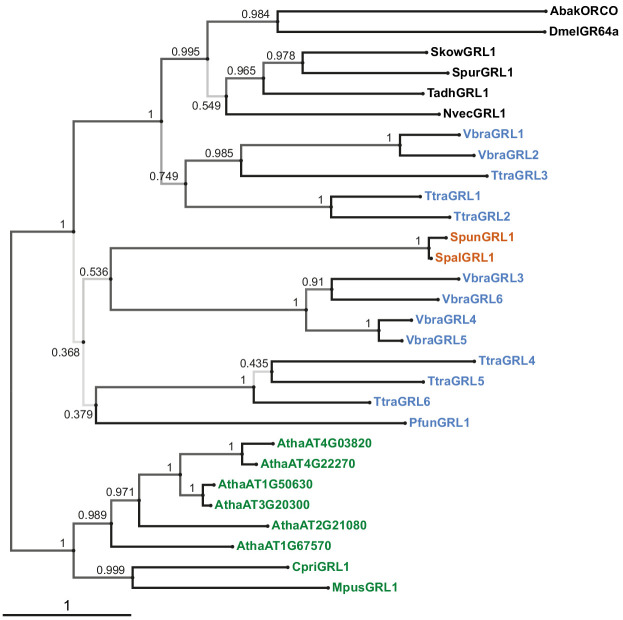
Figure 2. Conservation and divergence in TM7 features and GRL phylogeny.
(A) Side and top views of the cryo-EM structure of the ORCO homotetramer (Butterwick et al., 2018), in which the TM7 motif amino acid side chains are shown in stick format and colored red or orange. The region in the dashed blue box, representing the extracellular entrance to the ion channel pore, is shown in a magnified view on the far right. (B) Multiple sequence alignment of the C-terminal region (encompassing TM7) of unicellular eukaryotic GRLs and selected animal GRLs and plant DUF3537 proteins. Tadh, Trichoplax adhaerens; other species abbreviations are defined in Figure 1 and Table 1. The TM7 motif consensus amino acids (and conservative substitutions) are indicated below the alignment; h indicates a hydrophobic amino acid. Red dashed lines on the alignment indicate positions of predicted introns within the corresponding transcripts. Intron locations are generally conserved within sequences from different Kingdoms, but not between Kingdoms; many Protista sequences do not have introns in this region. (C) Maximum likelihood phylogenetic tree of unicellular eukaryotic GRLs, and selected animal GRLs and plant DUF3537 proteins, with aBayes branch support values. Although the tree is represented as rooted, the rooting is highly uncertain. Protein labels are in black for animals, orange for fungi, blue for protists and green for plants. The scale bar represents one substitution per site.