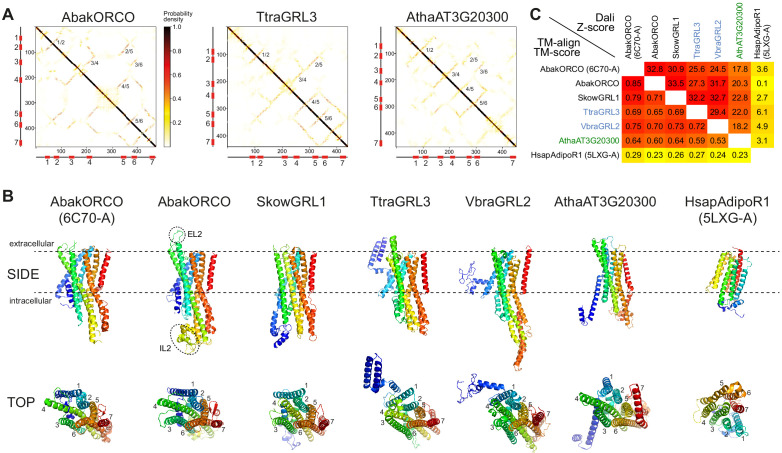
Figure 3. Ab initio structural predictions of GRLs and DUF3537 proteins.
(A) Inter-residue contact maps from trRosetta analysis of the indicated proteins. The axes represent the indices along the primary sequence; the positions of the predicted TM domains are shown in the schematics. The representation is mirror-symmetric along the diagonal; in one half ‘lines’ of contacts perpendicular to the diagonal of the map support the existence of anti-parallel alpha-helical transmembrane packing arrangements. Most pairs of predicted anti-parallel TMs are conserved across the proteins, despite variation in the length of loops between TM domains, supporting a globally similar packing of TM helices. The output of trRosetta analyses for these and other proteins is summarized in Supplementary file 7 and complete datasets are provided in the Dryad repository (doi:10.5061/dryad.s7h44j15f). (B) Side and top views of experimentally-determined (AbakORCO (PDB 6C70 chain A)) and Homo sapiens Adiponectin Receptor 1 (HsapAdipoR1; PDB 5LXG chain A [Vasiliauskaité-Brooks et al., 2017]) or the top trRosetta protein model of GRL and DUF3537 proteins. All GRL/DUF3537 proteins have a similar predicted global packing of TM domains (which is particularly evident in the top view in which the seven TM domains are labelled), despite variation in lengths of the loops and N-terminal regions (colored in dark blue). By contrast, HsapAdipoR1 has a fundamentally different arrangement of TM domains. The dashed ovals on the AbakORCO model highlight the extracellular loop 2 (EL2) and intracellular loop 2 (IL2) regions that were not visualized in the ORCO cryo-EM structure (Butterwick et al., 2018). (C) Quantitative pairwise comparisons of the structures shown in (B) using TM-align (Zhang and Skolnick, 2005) and Dali (Holm and Rosenström, 2010). TM-scores of 0.0–0.30 indicate random structural similarity; TM-scores of 0.5–1.00 indicate that the two proteins adopt generally the same fold (1.00 represents a perfect match). Dali Z-scores of <2 indicate spurious similarity. In both cases, these quantitative cut-offs are not stringent, and must be used as a guide in combination with other criteria (e.g., evidence for homology based upon primary sequence comparisons). The two half-matrices are colored using different scales.