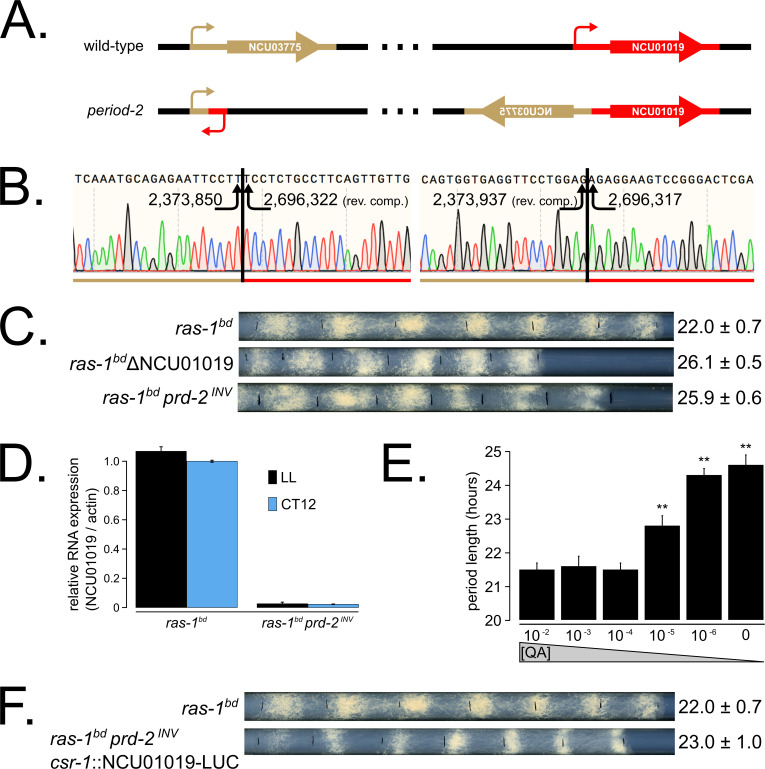
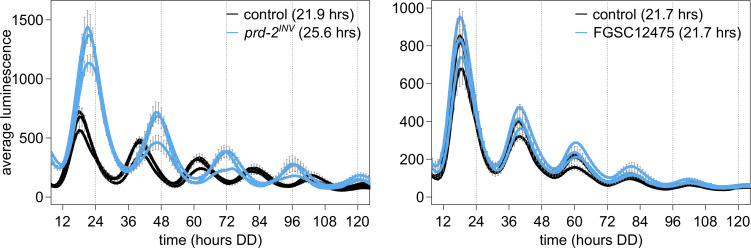
Figure 1. The prd-2 phenotype derives from reduced expression of NCU01019.
Whole genome sequencing identified a 322,386 bp inversion on linkage group V in the original prd-2 mutant strain (Lambreghts, 2012). The inversion breakpoints disrupt two loci, NCU03775 and NCU01019, depicted in cartoon form (A). Sanger sequencing confirms the DNA sequence of the left and right breakpoints, and the corresponding NC12 genome coordinates are shown at each arrowhead (B). Circadian period length was determined by race tube (RT) assay for ras-1bd controls, targeted deletion of the NCU01019 locus, and the classically derived prd-2INV mutant. The ΔNCU01019 mutant has a long period and slow growth defect similar to prd-2INV (C). NCU01019 RNA expression levels are detectable by RT-qPCR in the prd-2INV mutant but are drastically reduced compared to ras-1bd controls grown in constant light (LL) or at subjective dusk (CT12) during a circadian free run (D). After replacing the endogenous promoter of NCU01019 with the inducible qa-2 promoter, addition of high levels of quinic acid (10−2 to 10−3 M) led to a normal circadian period by RT assay (10−2 M τ = 21.5 ± 0.2 hr; 10−3 M τ = 21.6 ± 0.3 hr; 10−4 M τ = 21.5 ± 0.2 hr). Lower levels of QA inducer led to a long circadian period (10−5 M τ = 22.8 ± 0.3 hr; 10−6 M τ = 24.3 ± 0.2 hr; 0 QA τ = 24.6 ± 0.3 hr) due to reduced NCU01019 expression. Asterisks (**) indicate p<1 × 10−10 by Student’s t-test compared to 10−2 M QA RT results (E). The entire NCU01019 locus (plus 951 bases of its upstream promoter sequence) was fused in-frame with codon-optimized luciferase. Ectopic expression of this NCU01019-luc construct in the prd-2INV background rescues the long period phenotype by RT assay (F).