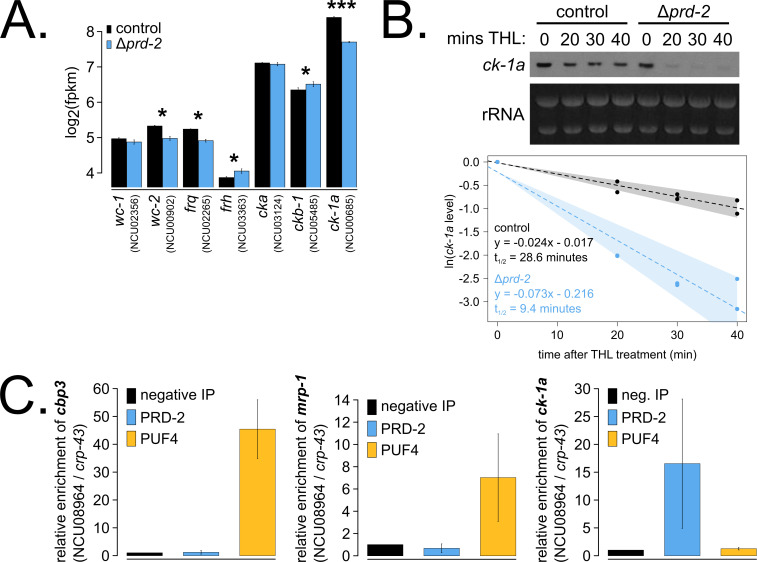
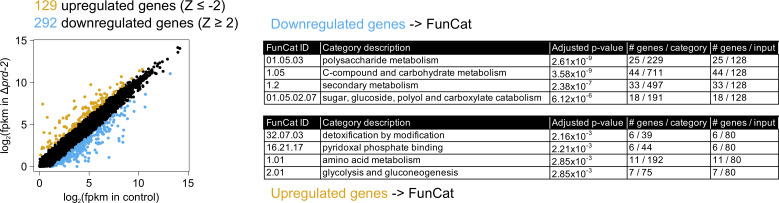
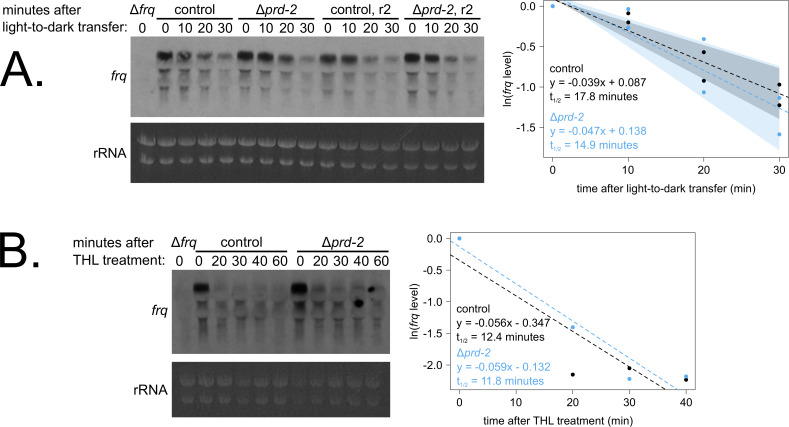
Figure 3. The core clock target of PRD-2 is the casein kinase I transcript.
Control and Δprd-2 cultures were grown in the light at 25°C in Bird medium for 48 hr prior to RNA isolation. Expression levels for core clock genes were measured by RNA-sequencing (N = 3 biological replicates per strain), and log2-transformed FPKM values are shown. Asterisks indicate p<0.05 (*) or p<5 × 10−5 (***) by Student’s t-test compared to control levels. The ck-1a transcript is >1.5× less abundant in Δprd-2 (A). ck-1a mRNA degradation kinetics were examined by Northern blot in a time course after treatment with thiolutin (THL) at approximately CT1 (N = 2 biological replicates). RNA levels were quantified using ImageJ, natural log transformed, fit with a linear model (glm in R, Gaussian family defaults), and half-life was calculated assuming first order decay kinetics (ln(2)/slope). Shaded areas around the linear fit represent 95% confidence intervals on the slope. The ck-1a transcript is 3× less stable in Δprd-2 (B). The PUF4 (NCU16560) RNA-binding protein pulls down known target transcripts cbp3 (NCU00057) and mrp-1 (NCU07386) by RT-qPCR (N = 3 biological replicates). PRD-2 CLIP samples were processed in parallel with PUF4 positive controls, and PRD-2 binds the ck-1a transcript in vivo (C).