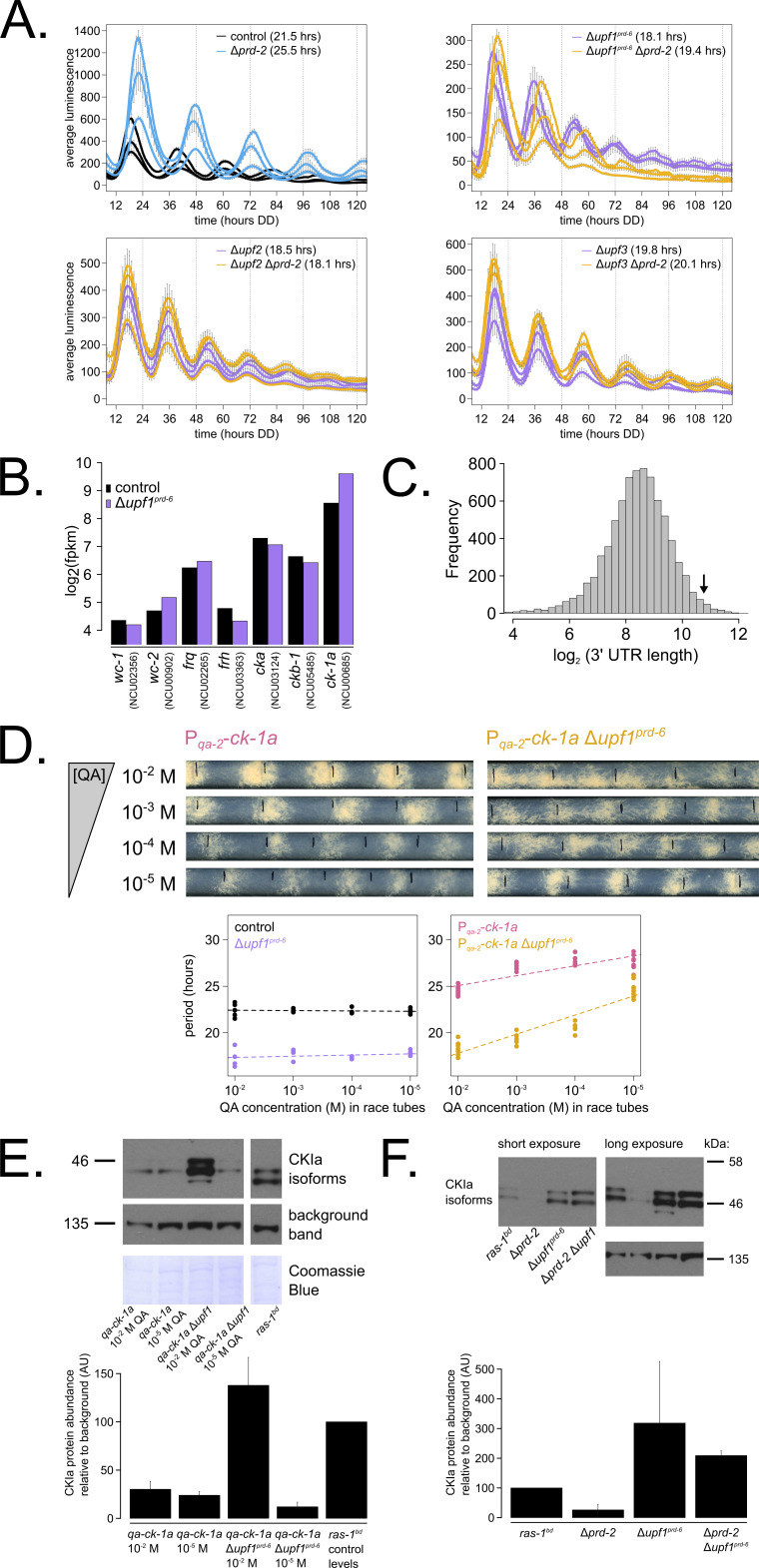
Figure 5. Nonsense-mediated decay (NMD) negatively regulates casein kinase I (CKI) levels via UPF1PRD-6, establishing a basis for the upf1prd-6 prd-2 genetic epistasis on circadian period length.
96-well plate luciferase assays were used to measure the circadian period length in triplicate wells per three biological replicate experiments for: ras-1bd controls (black, τ = 21.5 ± 0.3 hr), ras-1bd Δprd-2 (blue, τ = 25.5 ± 0.4 hr); ras-1bd Δupf1prd-6 (purple, τ = 18.1 ± 0.2 hr), ras-1bd Δupf1prd-6Δprd-2 double mutants (yellow, τ = 19.4 ± 0.7 hr); ras-1bd Δupf2 (purple, τ = 18.5 ± 0.5 hr), ras-1bd Δupf2 Δprd-2 double mutants (yellow, τ = 18.1 ± 0.3 hr); ras-1bd Δupf3 (purple, τ = 19.8 ± 0.3 hr), ras-1bd Δupf3 Δprd-2 double mutants (yellow, τ = 20.1 ± 0.2 hr). Each individual NMD subunit knockout is epistatic to the Δprd-2 long period phenotype (A). Raw RNA-seq data from a previous study (Wu et al., 2017) were analyzed using the same pipeline as data from Figure 3A (see Materials and methods). Control and Δupf1prd-6 gene expression levels (log2-transformed) are shown for core clock genes. The ck-1a transcript is >2× more abundant in Δupf1prd-6 (B). 3’-UTR lengths from 7793 genes were mined from the N. crassa OR74A genome annotation (FungiDB version 45, accessed on 10/25/2019), and plotted as a histogram. The arrow marks the 3’-UTR of ck-1a, which is 1739 bp and within the top 100 longest annotated UTRs in the entire genome (C). Representative race tubes (RTs) from ras-1bd Pqa-2-ck-1a single (pink) and ras-1bd Pqa-2-ck-1a Δupf1prd-6 double (yellow) mutants are shown at the indicated concentrations of quinic acid to drive expression of ck-1a. All results are shown in a scatterplot, where each dot represents one RT’s free running period length. ras-1bd controls (black) had an average period of 22.4 ± 0.4 hr (N = 20), and period length was not significantly affected by QA concentration (ANOVA p=0.605). ras-1bd Δupf1prd-6 controls (purple) had an average period of 17.5 ± 0.6 hr (N = 16), and period length was not significantly affected by QA concentration (ANOVA p=0.362). Period length of ras-1bd Pqa-2-ck-1a single mutants (pink) was significantly altered across QA levels (ANOVA p=2.9×10−8), and the average period at 10−5 M QA was 27.6 ± 0.8 hr (N = 8). Period length of ras-1bd Pqa-2-ck-1a Δupf1prd-6 double mutants (yellow) was also significantly affected by QA levels (ANOVA p=9.4×10−12), and the average period at 10−5 M QA was 24.7 ± 0.9 hr (N = 8). Thus, the double mutant period length was not genetically additive at low levels of QA induction, and the short period phenotype of Δupf1prd-6 is rescued (D). CKI protein levels were measured from the indicated genotypes grown in 0.1% glucose liquid culture medium (LCM) with QA supplemented at the indicated concentrations for 48 hr in constant light. A representative immunoblot of three biological replicates is shown, and replicates are quantified in the bar graph relative to ras-1bd control CKI levels from a 2% glucose LCM culture (E). CKI protein levels were measured from the indicated genotypes grown in 2% glucose LCM for 48 hr in constant light. A representative immunoblot of three biological replicates is shown, and replicates are quantified in the bar graph relative to ras-1bd control CKI levels (F). CKI protein levels are increased in Δupf1prd-6, decreased in the Δprd-2 mutant, and Δupf1prd-6 is epistatic to Δprd-2 with respect to CKI levels and circadian period length.