Figure 4.
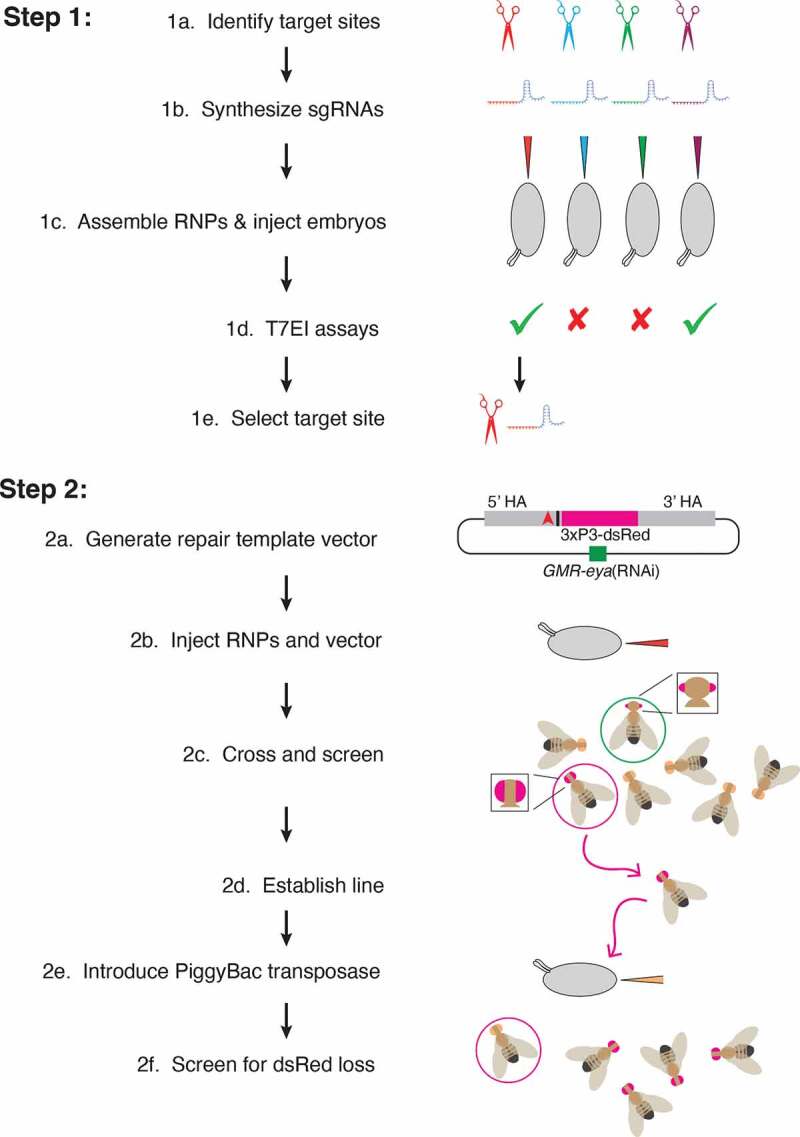
Workflow for two-step genome editing. (1a) Target sites flanking the area to be edited are identified (red, blue, green, purple) using online tools searching for optimal targets and with minimal off-target cleavage. (1b) Sequences from the selected target sites are transcribed in vitro to generate sgRNAs. (1c) Cas9 protein is incubated with sgRNAs before injection into embryos. (1d) Active sgRNAs that cleave embryo DNA are identified by T7 endonuclease I reactions. (1e) One of the active sgRNAs is chosen for genome editing in Step 2. (2a) Homology arms flanking the region of interest are cloned into the pBS-GMR-eya(shRNA) donor plasmid. In this example, the CRISPR target site (red triangle) is 5ʹ to the bases to be edited (black bar). (2b) Embryos are injected with the repair template plasmid and RNPs composed of sgRNA and Cas9 protein. (2c) Adult flies that develop from injected embryos are crossed back to the parental line. G1 progeny are screened for the DsRed marker. Positive G1 animals may have small eyes due to eya(shRNA) but these are not selected (green circle). Only positive G1 animals with normal eyes are selected (red circle). (2d) These are crossed to make purebred lines and molecularly analysed to determine if they contain the desired editing events. (2e) PiggyBac transposase is expressed in the germline, either by a single cross to a transgenic line, or in this example, by embryo injection of a plasmid expressing the transposase. (2f) Since the DsRed marker is dominant, adult flies developing from injected embryos that do not have red fluorescent eyes are then crossed and analysed with molecular tests to determine whether they have precisely excised the marker gene. Only the intended genomic edit remains
