Figure 4.
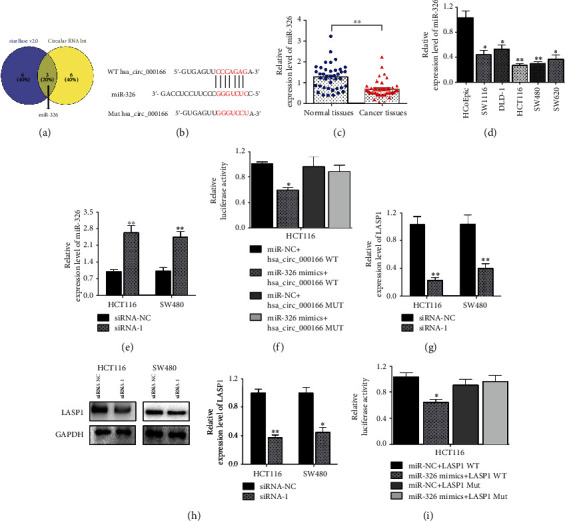
hsa_circRNA_000166 regulated CRC progression by targeting the miR-326/LASP1 axis. (a) The Venn analysis implied that 3 miRNAs, including miR-326, were involved in CRC. The blue represented 9 predicted miRNAs analyzed by starBase v2.0, and the yellow represented 9 predicted miRNAs analyzed by CircInteractome. (b) Binding site of hsa_circRNA_000166 in miR-326 was predicted by starBase. (c) qRT-PCR analysis showed that the transcriptional level of miR-326 was dramatically decreased in CRC tissues (n = 40) compared to the matched normal tissues (n = 40). (d) qRT-PCR analysis showed that miR-326 was notably downregulated in CRC cells compared with the normal colonic cells. (e) qRT-PCR assay displayed that miR-326 was significantly upregulated in siRNA-1-transfected groups compared with the controls in both HCT116 and SW480 cells. (f) Luciferase reporter assay indicated miR-326 dramatically repressed the WT hsa_circRNA_000166 luciferase activity but not Mut hsa_circRNA_000166 in HCT116 cells. (g, h) qRT-PCR and western blot assay showed that the transcriptional and translational levels of LASP1 were obviously downregulated in siRNA-1-transfected groups compared with the controls in both HCT116 and SW480 cells. (i) Luciferase reporter assay indicated miR-326 dramatically repressed the WT LASP1 luciferase activity but not Mut LASP1 in HCT116 cells. The Student t-test was used for statistics.
