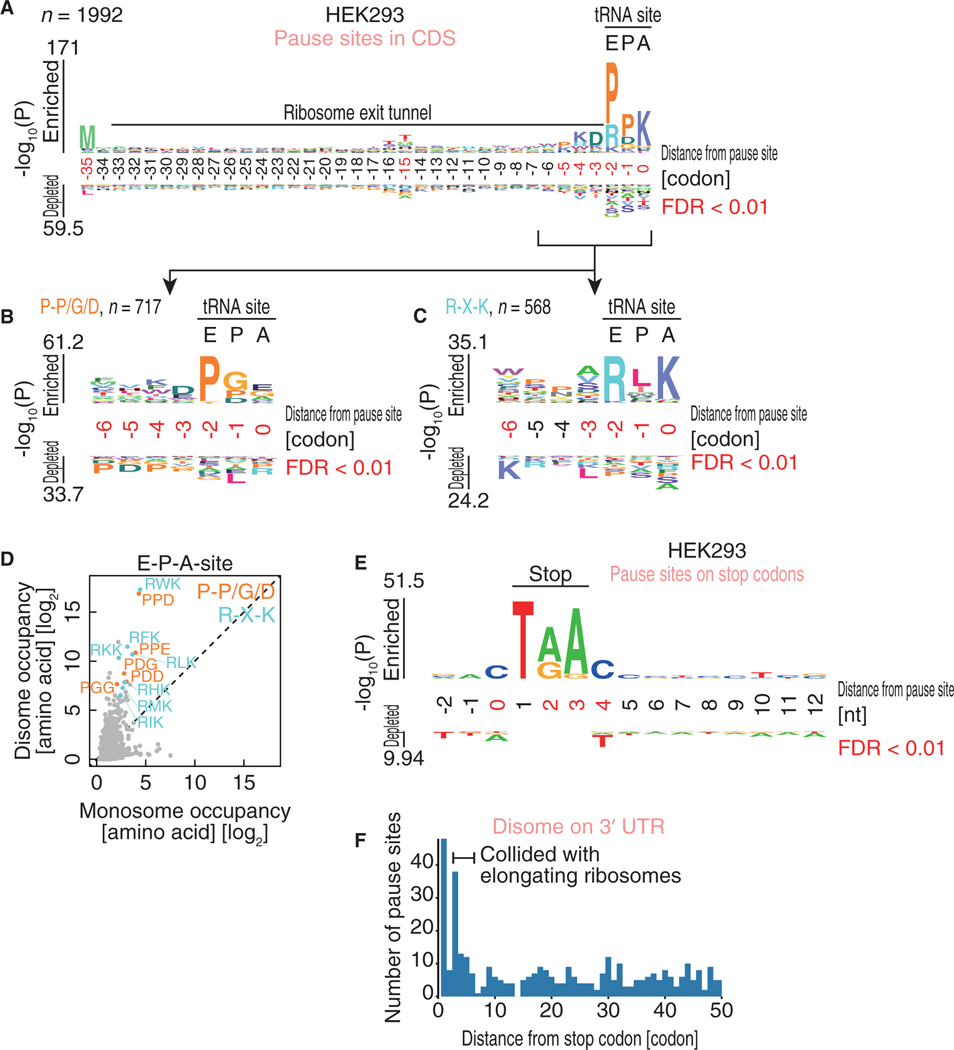
Figure 4. The Motifs Associated with Ribosome Collision.
(A–C) Amino acid sequences enriched in ribosome collision sites found in the CDS (A). The motifs are further sub-clustered (B and C). The codon position relative to the A-site in the leading ribosome is shown.
(D) Monosome and disome occupancies on tri-peptides at E-P-A-sites. Amino acids corresponding to Pro-Pro/Gly/Asp and Arg-X-Lys are highlighted in orange and light blue, respectively.
(E) Nucleic acid enrichment around ribosome collision sites on stop codons. Nucleic acids from the stop codon (where 1 indicates the first nucleotide in the stop codon) are depicted.
(F) Histogram of the number of disome pause sites found in the 3’ UTR along with the distance from the stop codon.
The logos in (A)–(C) and (E) were drawn with kpLogo (Wu and Bartel, 2017). The positions with significantly enriched sequences are highlighted in red.
See also Figure S4.