Fig. 2.
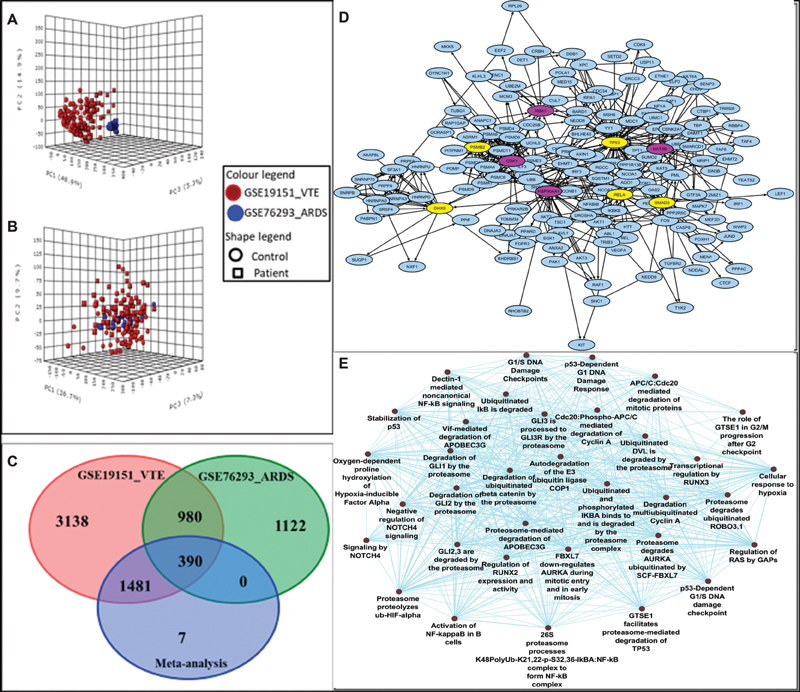
Result illustrations of comprehensive transcriptomics meta-analysis between acute respiratory distress syndrome (ARDS) and venous thromboembolism (VTE) using integrative meta-analysis of expression data (INMEX) tool ( A ) Plot of principal component analysis (PCA) as validation tool before batch effect removal and ( B ) after batch effect removal (using Combat method). ( C ) The Venn diagram comparing differentially expressed genes (DEGs) sets identified by the individual studies and by meta-analysis. The results obtained by the meta-analysis (1,878 DEGs) are compared with DEGs identified by individual study of VTE and ARDS. ( D ) Network depicting zero-order interaction of the shared DEGs in between ARDS and VTE datasets. Among the top 20 hub genes, few selected hub genes known to participate in host-viral interaction are shown in purple (overexpressed genes) and yellow (underexpressed gene) colors using Cytoscape. ( E ) Overrepresentation of pathways and gene ontology categories in biological networks identified from meta-analysis. Network representations of enriched pathway integrating the Kyoto Encyclopedia of Genes and Genomes (KEGG) and Reactome pathways along with Gene ontology for the selected top 20 hub genes using ClueGO Cytoscape plug-in. Hyper-geometric enrichment distribution tests, with an adjusted p -value of ≤0.05, followed by the Bonferroni adjustment for the terms and term groups were selected based on the highest significance.
