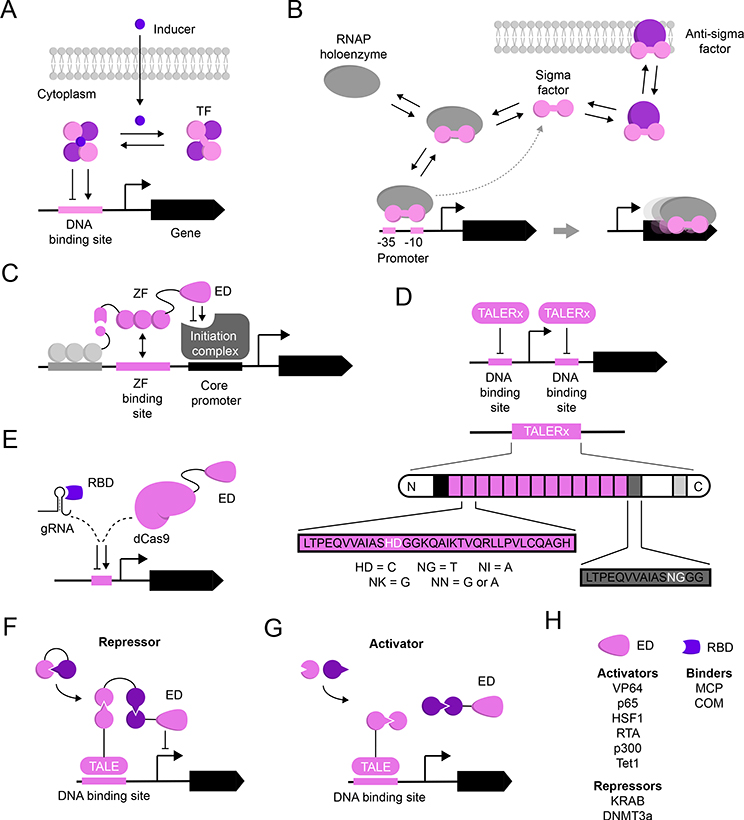
Fig. 3 |. Orthogonal Transcription Components.
Schematic representation of components fulfilling aspects of orthogonal transcription. Grayscale elements represent canonical or host-associated components. A) Small molecule-inducible transcription factors (TF): transcriptional units consist of a TF which binds to its cognate promoter in a small molecule-dependant manner [5]. B) Extracytoplasmic sigma (σ-factors): membrane-anchored anti-σ-factors bind their cognate σ-factor to inhibit association with the host RNAP holoenzyme. Following an external stimulus, the σ-factor dissociates from its anti-σ-factor, thereby allowing for σ-RNAP interaction and initiating transcription from its cognate promoter [54]. C) Zinc fingers (ZFs) are modular domains found in many eukaryotic TFs that make sequence-specific contacts with cognate DNA binding sites. Synthetic ZFs have been employed as programable TFs to define transcriptional states by fusing it with an effector domain (ED) [58]. D) TALERx is a programable TF platform based on transcription activator-like effectors (TALEs). Recognition is imparted by two residues in a repeating peptide sequences (pink rectangles with black outline) and correspond to specific nucleotide sequence in a designed DNA sequence [59]. E) Engineered CRISPR/dCas9-based transcriptional regulators: an engineered guide RNA (gRNA) enables dCas9 binding to the DNA sequence of interest. Either the protein (dCas9) or the gRNA can be modified to include additional functionality by recruiting trans-activating proteins [124]. F) De novo designed logic gates based on heterodimeric protein-protein interaction pairs [66]. These modular heterodimers can be used to generate transcriptional repressors (F) or activators (G). H) Effector domains and RNA-binding domains used to control gene expression [124].