Extended Data Figure 4 |. Single-nucleus RNA-sequencing (snRNA-seq) analysis of excitatory neuron clusters in the thalamus of Grn+/+ and Grn−/− mice.
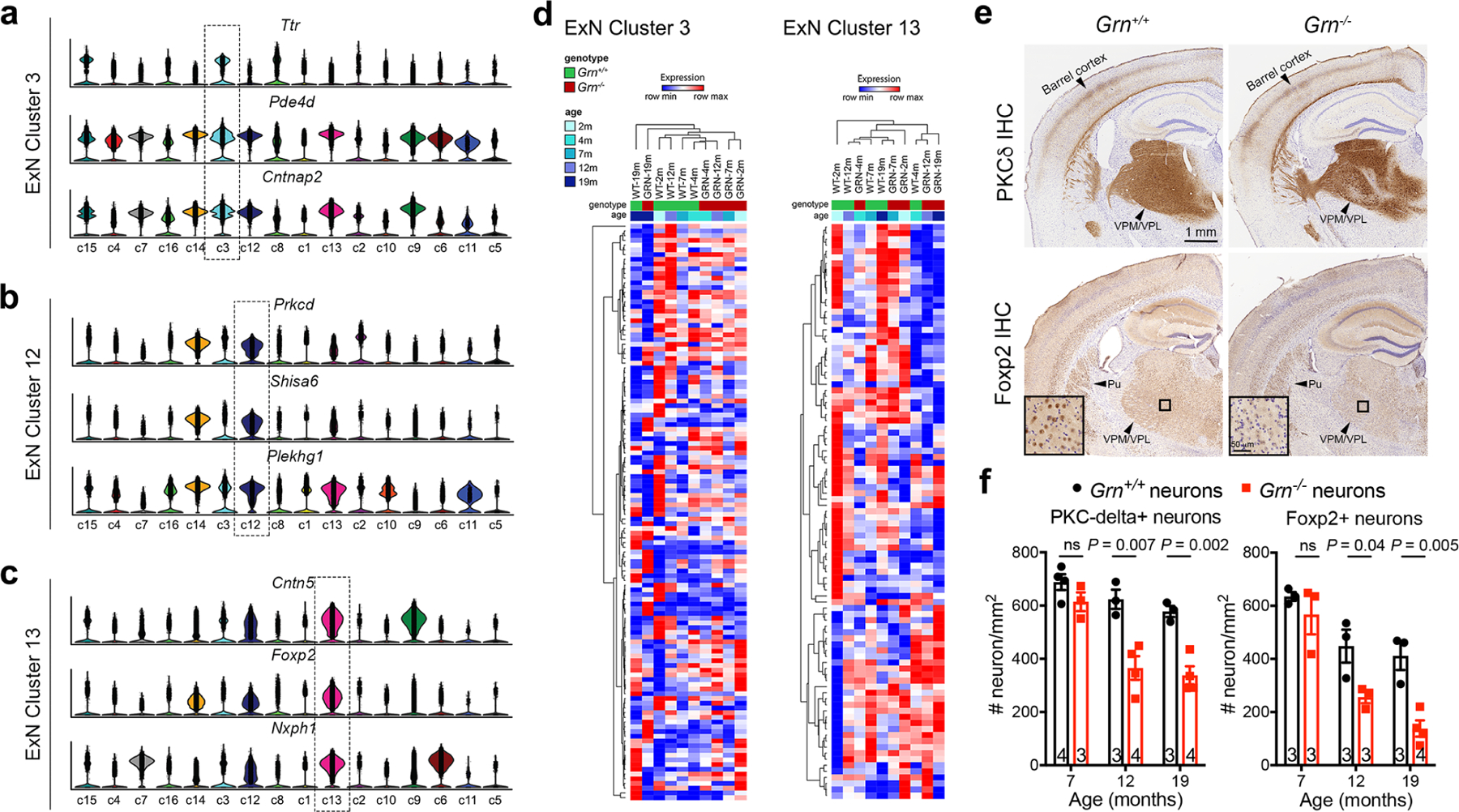
a–c. snRNA-seq identifies three distinct clusters of excitatory neurons based on the combined expression of Ttr (Transthyretin), Pde4d (Phosphodiesterase 4D) and Cntnap2 (Contactin associated protein like 2) in cluster 3, Prkcd (Protein kinase C Delta), Shisa6 (Shisa family member 6) and Plekhg1 (Pleckstrin homology and RhoGEF domain containing G1) in cluster 12, and Cntn5 (Contactin 5), Foxp2 (Forkhead Box P2) and Nxph1 (Neurexophilin 1) in cluster 13. d. Heatmaps of cluster 3 and cluster 13 show no definitive age-dependent changes in the transcriptomes. e. Comparison of excitatory neuron subtypes in 19 months old Grn+/+ and Grn−/− thalamus using immunohistochemical stains for PKCδ (upper panels) and Foxp2 (lower panels) reveals loss of PKCδ+ and Foxp2+ neurons, most prominently affecting neurons in the ventral posterolateral (VPL) and ventral posteromedial (VPM) nuclei of the thalamus. f. Stereology quantification of PKCδ+ and Foxp2+ neurons in the ventral thalamus of Grn+/+ and Grn−/− mice at 7, 12 and 19 months old. Data represent mean ± s.e.m., and the number of mice for each age and genotype is indicated at the bottom of each dataset. Statistics uses two-tailed unpaired Student’s t test. ns, not significant.
