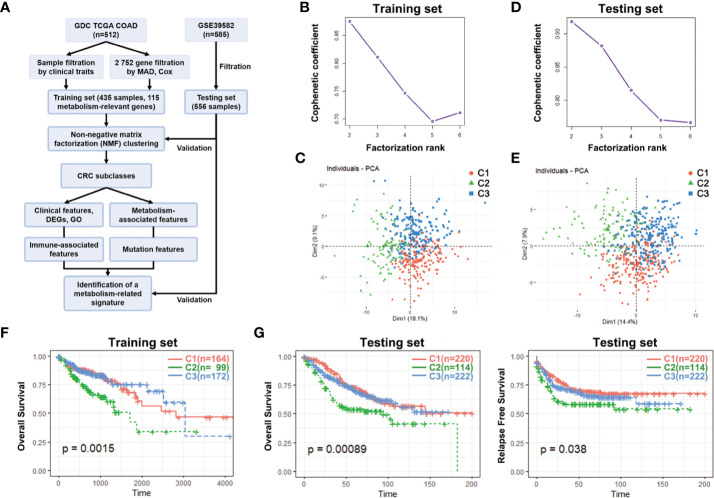
Figure 1.
Identification of CRC subclasses using NMF consensus clustering. (A) A flow chart of the study. (B) NMF clustering using 115 metabolism-associated genes in training set. Cophenetic correlation coefficient for k = 2–6 is shown. (C) PCA showed the distribution of three CRC subclasses in training set. (D) Cophenetic correlation coefficient in testing set. (E) The distribution of three CRC subclasses in testing set. (F) OS of three subclasses (C1, C2, and C3) in training set. (G) OS and RFS of three subclasses in testing set. CRC, Colorectal cancer; NMF, Non-negative matrix factorization; MAD, Median absolute deviation; PCA, Principal components analysis; OS, Overall survival; RFS, Relapse free survival; DEGs, Differentially expressed genes; GO, gene ontology.