Figure 1. mRNA accumulation of SNARE and vesicle trafficking associated transcripts following On-lz infection of resistant and susceptible tomato varieties.
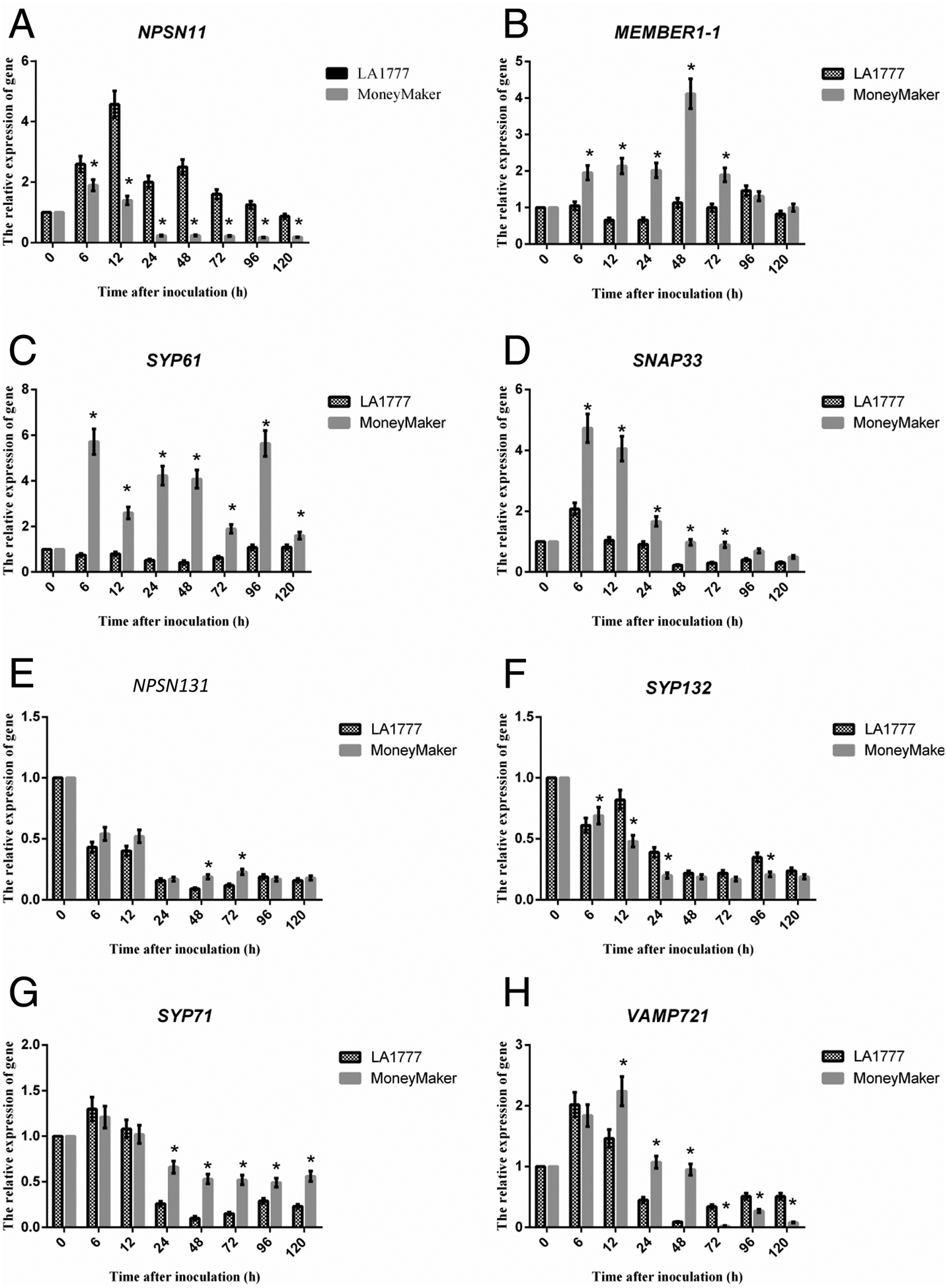
Quantitative real-time PCR (qRT-PCR) analysis of mRNA accumulation of SNAREs genes. (A) NPSN11 was significantly up-regulated in resistant LA1777 than susceptible cultivar Money Maker. In contrast, (B) MEMBER1–1, (C) SYP61, and (D) SNAP33 were significantly up-regulated in susceptible cultivar Money Maker than resistant LA1777. Meanwhile, (E) NPSN13, (F) SYP132, (G) SYP71, and (H) VAMP721 were also significantly up-regulated in susceptible cultivar Money Maker than resistant LA1777 at the end of the test time. For mRNA expression analyses, 4-week-old LA1777 (resistant) and Money Maker (susceptible) tomato plants (leaves) were spray inoculated with On-lz (~105 spores ml−1) and incubated at 22°C for up to 5 days. For analysis of mRNA accumulation following pathogen infection, samples were collected at the indicated time points (time after-inoculation (h)). Total RNA was extracted from leaves and one microgram of total RNA was used for first-strand cDNA synthesis. All DNA primers used for quantitative real-time PCR (qPCR) are listed in Supplementary Table S1. GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE (SlGAPDH) was used as an internal control for amplification. Expression values are represented as mean ± standard error of the mean (SEM). Statistical analysis was evaluated using a two-way ANOVA, followed by the Bonferroni post-test as compared with time 0. P values ≤0.05 were considered significant, where * P < 0.05.
