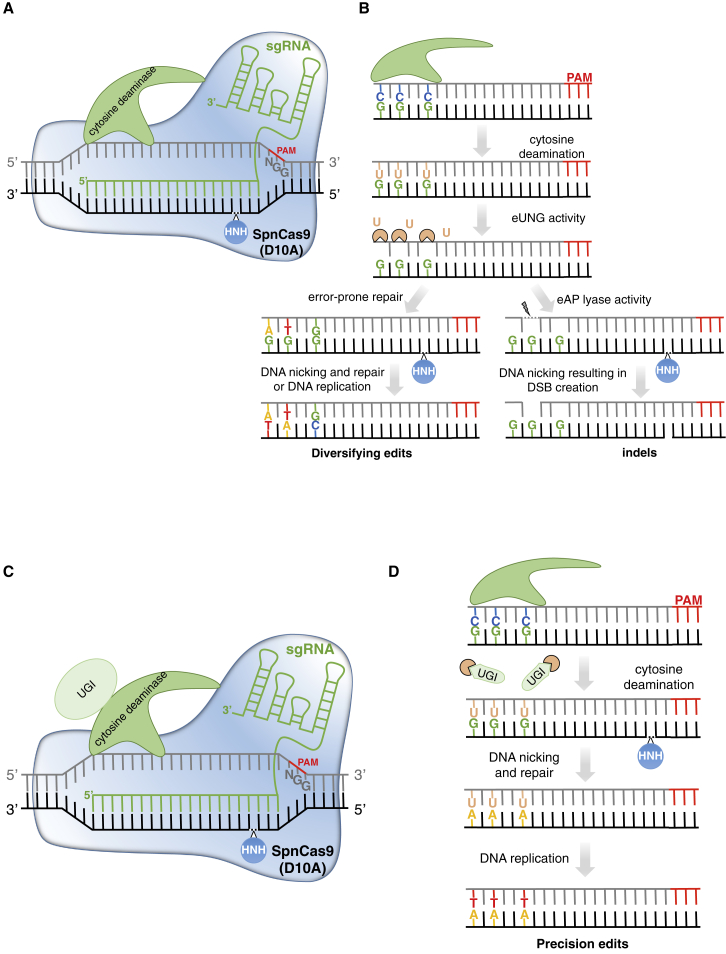
Figure 3.
CRISPR-Mediated Base Editing Using CBEs.
(A) CBEs are composed of nCas9 (D10A) fused to a cytosine deaminase catalytic domain (rAPOBEC1, PmCDA1, hAID, or hA3A) that mediates cytosine deamination in the so-called editing window at the 5′ end of the non-targeted sequence.
(B) After C deamination into U, endogenous uracil DNA glycosylase (eUNG) detects and removes the U, leading to an abasic site, which is further processed through error-free (U-to-C) or error-prone repair, producing different base substitutions, albeit at the cost of indel mutations due to the generation of DSBs through concomitant ssDNA breaks by nCas9 and endogenous AP lyases (eAP lyase). This system allows the production of C-to-T, C-to-G, and C-to-A conversions.
(C) CBE architecture can be upgraded through the fusion of one to several uracil glycosylase inhibitors (UGIs) to the base editor to increase the rate of C-to-T conversion while limiting the formation of by-products.
(D) After C deamination, UGIs protect the U edits from eUNG, thereby preventing the formation of abasic sites and mostly producing C-to-T conversions through the nicking of the non-edited strand and the intervention DNA repair/replication mechanisms, with a low level of by-products, such as indel mutations.
The schemes are not at scale and are for illustrative purposes only.