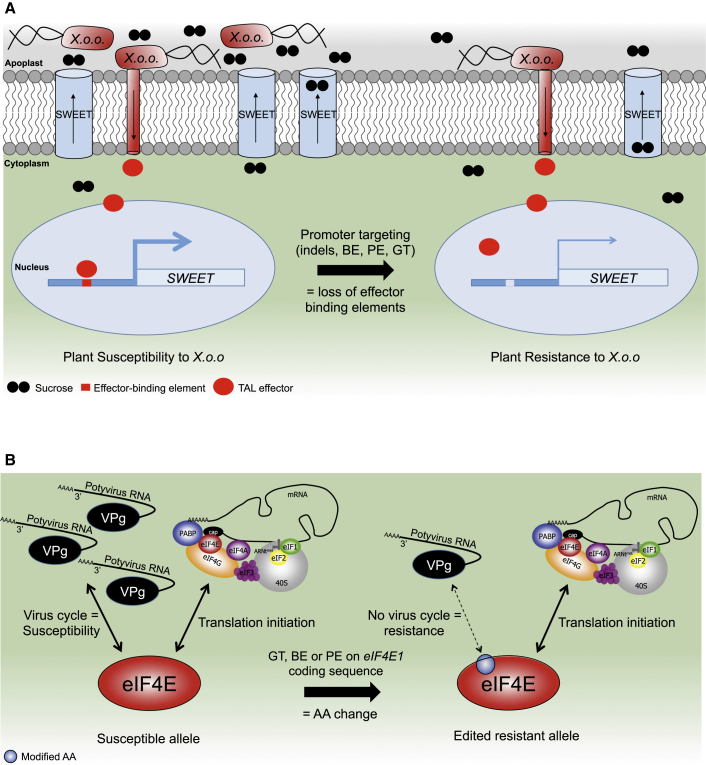
Figure 7.
Representative Model of Editing Resistance by Loss of Susceptibility.
(A) Resistance to bacteria through editing the SWEET promoter. During infection leading to susceptibility (left side), Xanthomonas oryzae pv. oryzae (Xoo) bacteria express transcription activator-like effectors (TAL effector) in the plant cell. These effectors bind effector-binding elements (EBE) located in the promoters of the SWEET genes that encode sucrose transporters. The binding triggers the activation of SWEET genes, and of the encoded sucrose transporter, and results in an increase in sucrose content in the apoplast. The excess of sucrose benefits to the bacteria and contribute to its multiplication. Genetic resistance can be engineered (right side) by removing the EBE region(s) from the SWEET promoter region: the SWEET gene is no longer activated by the TAL effector, such that sucrose content stays low in the apoplast, resulting in resistance.
(B) Resistance to potyvirus through base editing of the translation initiation factor eIF4E. In susceptible plants (left side), the translation initiator eIF4E is necessary for the infection cycle of potyviruses, represented by their ssRNA+ genome linked in 5′ to the viral protein genome-linked (VPg). At the same time, eIF4E proteins are involved in translation initiation of the host mRNA for protein synthesis. Base editing of the eIF4E coding sequence (right side) can be used to introduce non-synonymous mutations associated with amino acid changes usually found in resistance alleles from the natural diversity of plants. This mutation does not affect the translation initiation function of eIF4E while suppressing its interaction with potyvirus, leading to resistance. This allows resistance enhancement at no developmental cost. The translation initiation complex depiction is adapted from Robaglia and Caranta (2006).
The schemes are not at scale and are for illustrative purposes only.