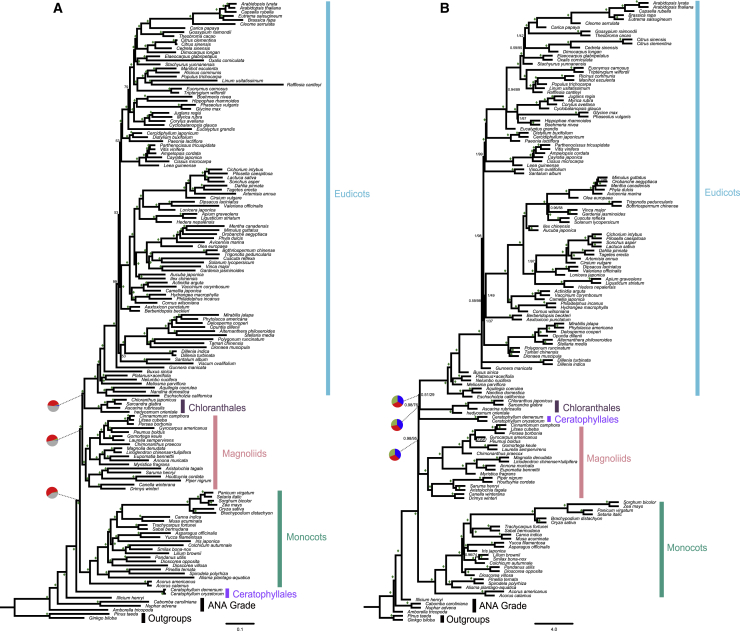
Figure 2.
The Species Trees Inferred from the Dataset of 1594 Nuclear Gene.
(A) The concatenation-based species tree inferred by IQ-TREE. Numbers associated with nodes are the ultrafast bootstrapping support (UFboot); diamonds indicate UFboot support of 100%. The branch lengths in substitutions per site were estimated by IQ-TREE. The pie charts at three backbone nodes of angiosperms present the proportion of gene tree concordance and conflict in the 1594-gene dataset. Pie chart color coding: blue, fraction of gene trees that are concordant with the species tree; green, fraction of gene trees supporting the second most common conflicting topology; red, fraction of gene trees supporting all other alternative conflicting partitions; gray, fraction of gene trees with <50% bootstrap support at that node.
(B) The coalescent-based species tree was inferred by ASTRAL. Numbers associated with nodes are support values obtained by the posterior probability (PP, on the left) and multilocus bootstrapping (MLBS, on the right). Diamonds indicate PP of 1.0 and MLBS of 100%. The branch lengths in coalescent units were estimated by ASTRAL. Pie charts show relative quartet support for the current tree (blue) and the two (green and red) alternative quartets.