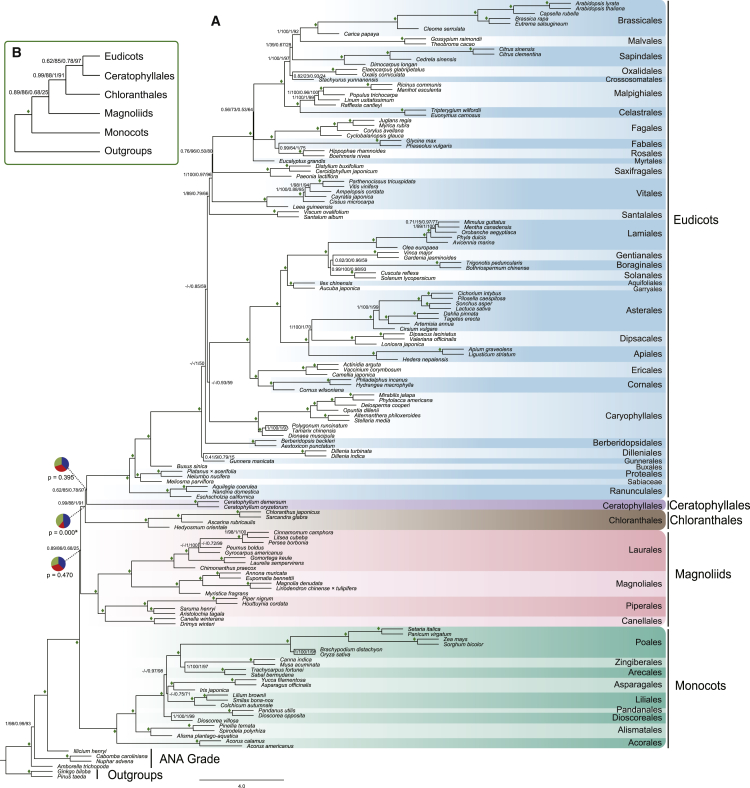
Figure 3.
The Coalescent-Based Species Tree of Angiosperms Is Supported by the Trees Inferred from Two Subsets of Nuclear Genes.
(A) The tree inferred by ASTRAL using 296 genes. The detailed phylogeny of 756 genes is presented in Supplemental Figure 4. Numbers associated with nodes are support values: posterior probability and multilocus bootstrapping (PP and MLBS). The first two are PP and MLBS values from 756 genes, and the last two are PP and MLBS values from 296 genes. Diamonds indicate PP of 1.0 and MLBS of 100% in both datasets. The branch lengths are in coalescent units, as estimated by ASTRAL. The pie charts at three backbone nodes of angiosperms show relative quartet support for the current tree (blue) and the two (green and red) alternative quartets. p values for the polytomy test are given for four backbone nodes of angiosperm below the respective pie charts for those nodes, and significance (p ≤ 0.05) is indicated with an asterisk.
(B) An abbreviated tree showing the relationship of the five lineages of Mesangiospermae with associated support values (PP/MLBS).