Figure 1.
Advantages and Experimental Procedure for Quantitative Abundance Profiling of the Microbiome in Plant Roots.
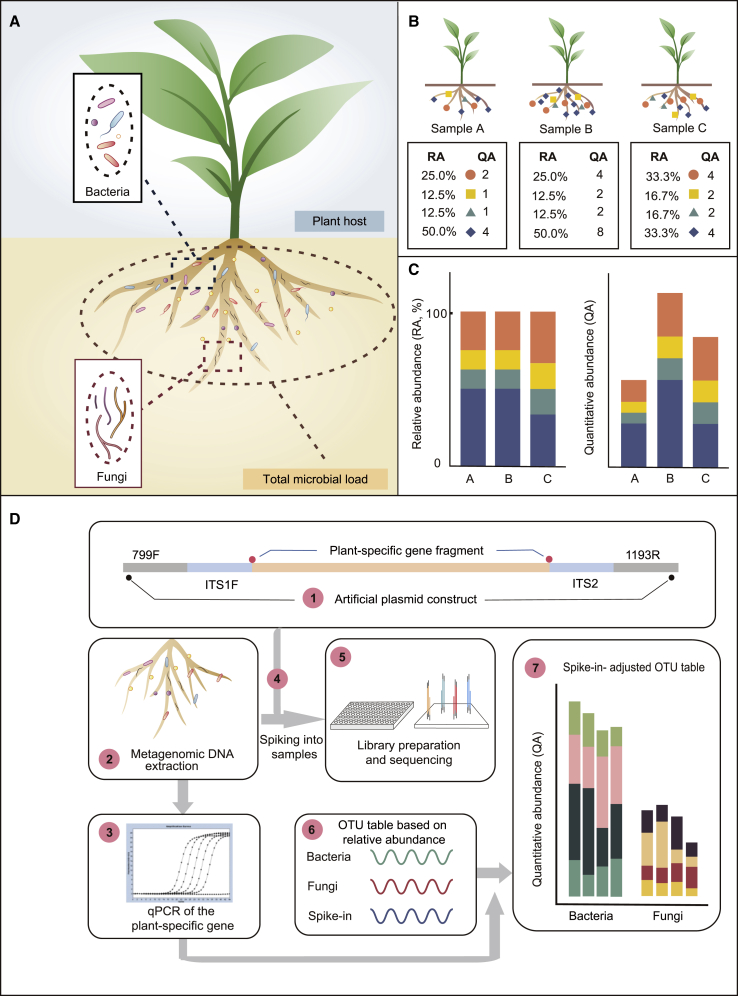
(A) The major limitation of the current root microbiome profiling technique based on relative abundance is the lack of a method for quantitatively assessing the microbial load per unit amount of plant root tissue.
(B) Three simulated root microbiomes associated with the same amounts of root tissue (upper panel) and the corresponding microbial profiling based on relative or quantitative abundance (lower panel). RA, relative abundance; QA, quantitative abundance, representing the copy-number ratio of microbial marker genes relative to plant genome.
(C) Bar plots showing a comparison of relative (left panel) versus quantitative (right panel) microbial profiling of simulated root microbiome samples in (B).
(D) Schematic diagram showing the workflow of HA-QAP. (1) The artificial plasmid is designed and constructed as a spike-in control, containing conserved 16S rRNA, ITS primer regions (799F/1193R and ITS1F/ITS2), and a plant-specific gene sequence (the RID1 gene in this study). (2) DNA is extracted from the root microbiome, including plants and microbes. (3) A selected plant marker gene found in the total DNA is quantified by qPCR; this value is used to calibrate microbial relative abundance to quantitative abundance in step (7). (4) The predefined amount of spike-in plasmid is added to the DNA of root microbiome samples based on experience. (5 and 6) PCR amplicon libraries (bacteria and fungi, respectively) are prepared, sequenced (5), and analyzed (6). (7) The bacterial and fungal reads are calibrated based on the read counts of the spike-in and the amount of the plant marker gene quantified by qPCR in step (3) to determine the quantitative abundance relative to the host plant tissue.