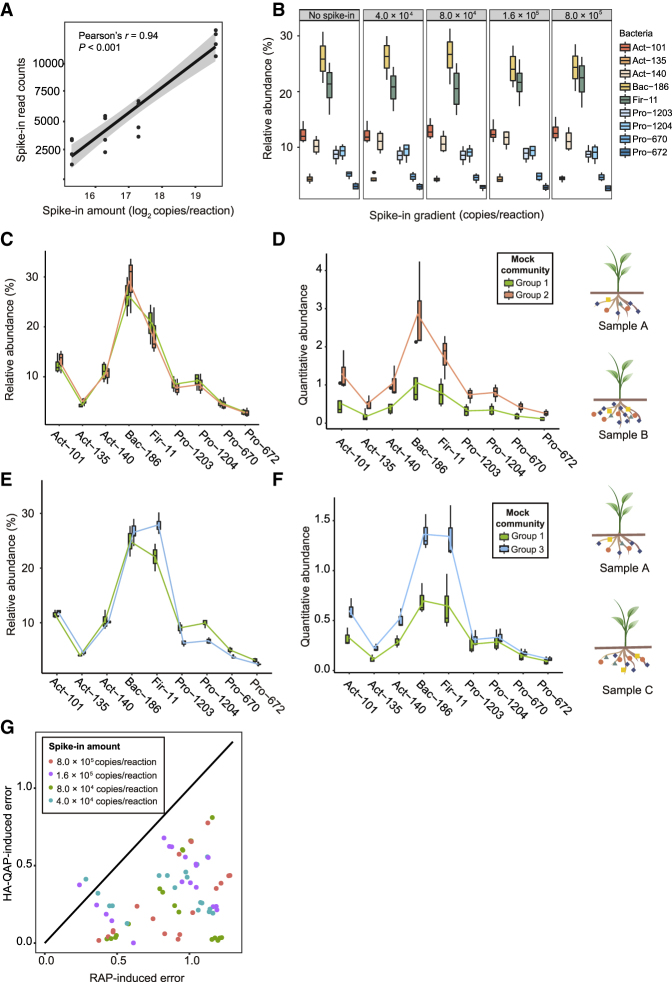
Figure 2.
HA-QAP Is More Accurate than Classical Profiling Based on Relative Abundance in the Bacterial Mock Experiments.
(A) Dose–response curves for the linear correlation between read counts of spike-in plasmid (BI12-4) obtained by Illumina sequencing and amount of spike-in plasmid in the DNA samples, indicating that bacterial reads of the spike-in plasmid in the sequencing data reflect the amount of spike-in plasmid in the initial DNA samples. The gray region indicates 95% confidence intervals (CIs).
(B) Box plots representing the relative abundance of the nine bacteria in the same mock experiment with a gradient of spike-in levels of 0–8.0 × 105 copies per reaction. Wilcoxon rank-sum test showed no significant differences in bacterial relative abundance between the control group without spike-in and groups with different spike-in levels (4.0 × 104, 8.0 × 104, 1.6 × 105, and 8.0 × 105 copies per reaction, P > 0.05).
(C and D) The HA-QAP method revealed the significant increase in bacterial load (two-sided t-test, P < 0.05), which could not be detected by the classical method based on relative abundance. Box plots showing a comparison of bacterial profiles in mock experiments between groups 1 and 2 using the classical method based on relative abundance (RAP) (C) and HA-QAP (D). Quantitative abundance represents the copy-number ratio of bacterial 16S rRNA genes relative to plant genome.
(E and F) The HA-QAP method improved the detection of the increases in the levels of Actinobacteria, Bacteroidetes, and Firmicutes (two-sided t-test, P < 0.05) and revealed that the quantitative abundance of Proteobacteria did not change (two-sided t-test, P > 0.05) when the amounts of other bacteria increased. Box plots showing a comparison of bacterial profiles in mock experiments between groups 1 and 3 using RAP (E) and HA-QAP (F). Quantitative abundance represents the copy-number ratio of bacterial 16S rRNA genes relative to plant genome. Notably, the RAP method showed a spurious reduction in Proteobacteria levels, but HA-QAP did not.
(G) Scatter plot showing the ratio of errors between HA-QAP and RAP. Dots on the line with fixed slope = 1 represent errors from HA-QAP equivalent to those from RAP. Most dots fall below the line with slope = 1, demonstrating that the HA-QAP method presents more real data. Data are based on comparisons (group 1 versus group 2; group 1 versus group 3) at different spike-in levels. Three groups of mock experiments were designed; n = 3, 4, and 5 for groups 1, 2, and 3, respectively.
All data shown in (C) to (F) are from samples with 4.0 × 104 copies of spike-in per reaction. The trend was consistent using other amounts of spike-in. Act, Actinobacteria; Bac, Bacteroidetes; Fir, Firmicutes; Pro, Proteobacteria. See also Supplemental Figures 2 and 3.