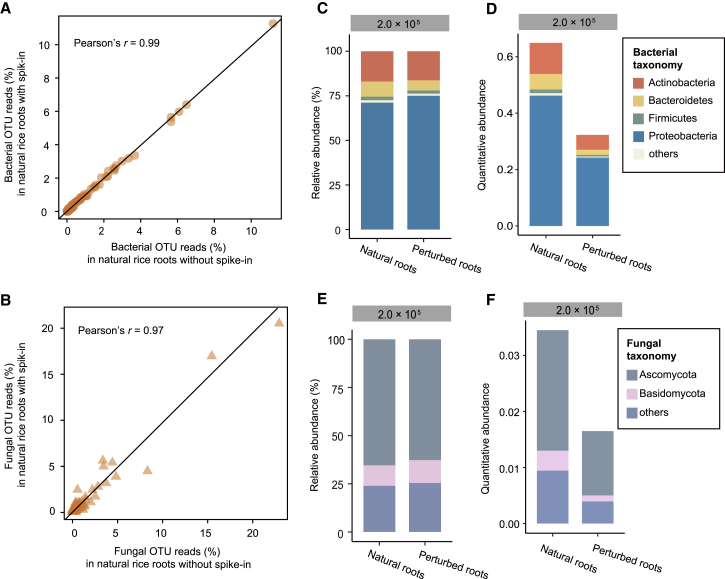
Figure 4.
HA-QAP Reveals a Reduction in Microbial Load in Perturbed Natural Root Samples.
(A and B) Scatter plot showing the correlation between the relative abundance of bacterial OTUs (A) and fungal OTUs (B) in natural rice roots with spike-in (spike-in levels: 2.0 × 105 copies per reaction, in y axis) versus rice roots without spike-in (x axis), demonstrating that spike-in did not influence the relative abundance of bacterial or fungal OTUs in natural root samples.
(C and D) Bacterial profile based on relative abundance (C) and quantitative abundance (D) in natural rice roots and rice roots with 55% microbial load compared with the original natural samples by RAP and HA-QAP, respectively. Quantitative abundance represents the copy-number ratio of bacterial 16S rRNA genes relative to plant genome. Note that the HA-QAP method revealed the reduction in bacterial load in perturbed natural rice roots.
(E and F) Fungal profile based on relative abundance (E) and quantitative abundance (F) in natural rice roots and rice roots with 55% microbial load compared with the original natural samples by RAP and HA-QAP, respectively. Quantitative abundance represents the copy-number ratio of fungal ITS relative to plant genome. Note that the HA-QAP method revealed the reduction in fungal load in perturbed natural rice roots.
Data in (C) to (F) represent three technical replicates.