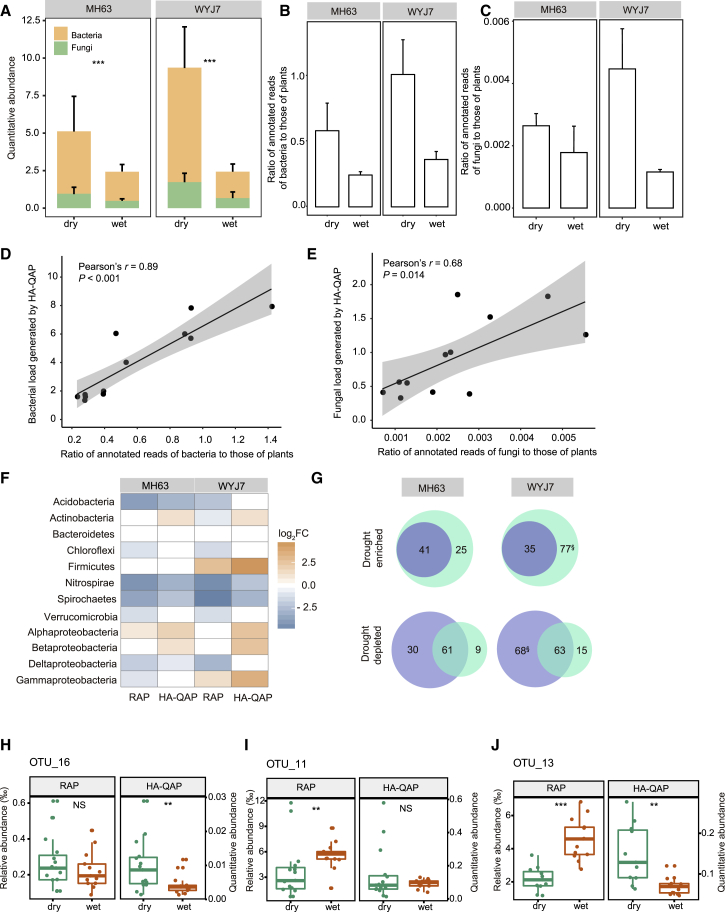
Figure 5.
HA-QAP Reveals Microbial Load Increase in Rice Roots under Drought Stress.
(A) The HA-QAP method showing the significant increases in bacterial and fungal loads in the root microbiome of two rice varieties (MH63 and WYJ7) under drought-stress conditions. Quantitative abundance represents the copy-number ratio of bacterial 16S rRNA genes and fungal ITS relative to plant genome.
(B and C) Metagenomic data showing higher ratios of annotated reads of bacteria (B) and fungi (C) to those of plants under drought conditions in rice varieties MH63 and WYJ7 grown in Hainan (dry: n = 3; wet: n = 3).
(D and E) Scatter plots showing the linear correlation between the ratio of annotated reads of bacteria (D) and fungi (E) to those of plants detected by metagenomic sequencing (x axis) versus the microbial load detected by the HA-QAP method (y axis). Gray region indicates 95% CIs.
(F) Heatmap showing the log2 fold change in abundance of drought-responsive bacterial phyla with respect to control treatment (wet) in rice varieties MH63 and WYJ7 detected by HA-QAP and RAP. Orange boxes indicate an increase while blue boxes indicate a decrease. White boxes indicate no significant fold change. Note that for the root microbiome of both varieties, the HA-QAP method revealed a shift in drought-responsive phyla, with an enhanced enrichment and reduced depletion under drought-stress conditions compared with wet conditions due to the increase in root bacterial load detected by HA-QAP.
(G) Venn diagram showing the overlap and differences in drought-responsive OTUs detected by the RAP and HA-QAP methods. The purple and green ellipses represent drought-responsive OTUs detected by the RAP and HA-QAP method, respectively. § indicates the five OTUs detected as depleted OTUs using RAP but identified as enriched OTUs using HA-QAP, suggesting that the microbial load value is essential for tracking the changes in the abundances of these bacteria.
(H–J) Examples showing the changes in OTUs in rice roots under drought and wet conditions using RAP and HA-QAP. Quantitative abundance represents the copy-number ratio of bacterial 16S rRNA genes relative to plant genome. The responses of three OTUs to drought-stress conditions differed depending on the method: OTU_16 (H) was detected as an enriched group responsive to drought only using HA-QAP. Conversely, OTU_11 (I) was detected as significantly depleted in dry root samples only using RAP and not HA-QAP. OTU_13 (J) was found to be significantly depleted in dry root samples using RAP but enriched using HA-QAP.
Note that all data from Hainan are consistent with those from Anhui (see also Supplemental Figure 9 and 10). For all analyses (except for B and C), the number of biological replicates was as follows: MH63 (dry: n = 15; wet: n = 13); WYJ7 (dry: n = 11, wet: n = 13); bulk soil (dry: n = 6; wet: n = 6). Error bars represent SD calculated from replicates. Statistical significance was determined by Wilcoxon rank-sum test. Asterisks indicate statistically significant differences (**P < 0.01; ***P < 0.001). NS, not significant.